Molecular characterization of two microalgal strains in Egypt and investigation of the antimicrobial activity of their extracts
Helwan University. Faculty of Science. Department of Botany and Microbiology. Helwan (Egypt). E-mail: nerminel_semary@yahoo.co.uk
National Organization for Drug Control. Al Harram. Giza (Egypt).
Received on April 12, 2012 ; accepted on April 15, 2013.
Résumé
Caractérisation moléculaire de deux souches de micro-algues issues d’Égypte et recherche d’une activité antimicrobienne de leurs extraits. L’émergence de nouveaux agents pathogènes et l’augmentation de la résistance aux médicaments de ceux qui sont connus posent un défi difficile. Des composés bioactifs, économiquement rentables, récemment découverts, sont étudiés à cette fin. Des micro-algues en constituent une source prometteuse. Dans le présent travail de recherche, nous avons pu isoler et identifier deux souches de micro-algues en utilisant l’analyse moléculaire et phylogénétique et nous montrons qu’une des deux souches appartient aux chrysophycées et l’autre aux cyanobactéries. Nous rapportons également une activité antimicrobienne de certains de leurs extraits lipophiles. Plusieurs fractions ont montré un niveau élevé de bioactivité antimicrobienne contre plusieurs organismes résistants à plusieurs médicaments, Salmonella sp., Citrobacter sp., Aspergillus niger et Aspergillus flavus. La fraction 3 de Poterioochromonas malhamensis a montré le niveau d’activité le plus élevé contre deux pathogènes bactériens résistants à plusieurs médicaments. Le diamètre de la zone d’inhibition était de 1,4 cm pour Salmonella et 1,4 cm pour Citrobacter. Parallèlement, une autre fraction lipophile de la cyanobactérie Synechocystis salina a présenté une activité biologique à large spectre (le diamètre de la zone d’inhibition était de 0,9 cm pour Aspergillus niger, 1 cm pour Citrobacter et 0,9 cm pour Salmonella). Une fraction lipophile d’Aphanizomenon a montré une bioactivité antifongique contre Aspergillus niger et Aspergillus flavus, les zones d’inhibition ayant un diamètre de 1,1 cm et 1,0 cm, respectivement. Cette étude met en évidence l’activité antimicrobienne de micro-algues locales et démontre l’intérêt de la mise sur pied de programmes de dépistage de ces micro-organismes.
Abstract
The emergence of new pathogens and the increasing drug-resistance of recognized ones pose a difficult challenge. One way that this challenge is being addressed is through the discovery of new cost-effective drug resources in the form of bioactive compounds. Algae represent a promising source of bioactive compounds in this regard. In the present research, we used molecular and phylogenetic analysis to isolate and identify two microalgal strains. We found that one strain belonged to the phylum chrysophyta and the other to the cyanobacteria. We also investigated the antimicrobial activity of some of the lipophilic extracts of the two microalgal strains. Several fractions showed high individual antimicrobial bioactivity against multidrug-resistant Salmonella sp., Citrobacter sp., Aspergillus niger and Aspergillus flavus. Fraction III from Poterioochromonas malhamensis showed the highest level of activity against two multidrug-resistant bacterial pathogens. The inhibition zone diameter was 1.4 cm for Salmonella and 1.4 cm for Citrobacter. Meanwhile, another lipophilic fraction from the cyanobacterium Synechocystis salina showed broad-spectrum bioactivity (inhibition zone diameter of 0.9 cm for Aspergillus niger, 1 cm for Citrobacter and 0.9 cm for Salmonella). One lipophilic fraction from Aphanizomenon showed antifungal bioactivity against Aspergillus niger and Aspergillus flavus, where the inhibition zone diameter was 1.1 cm and 1.0 cm, respectively. The study highlights the antimicrobial bioactivity of extracts from local microalgae and emphasizes the importance of carrying out screening programs for those microorganisms.
1. Introduction
1Algae are oxygenic phototrophs found in all water bodies worldwide. They comprise different groups of organisms, varying widely in their morphology, anatomy and habitats and capable of surviving in the most extreme niches (Whitton et al., 2000). Microalgae have the ability to produce a wide range of bioactive compounds (Thajuddin et al., 2005), enabling them to cope with their environments and to out-compete other co-existing microorganisms through the antimicrobial effect of those compounds (Mundt et al., 2003). It is well known that the current sources of antimicrobial compounds mostly originate from actinomycetes and plants (Borowitzka, 1995). We are currently witnessing the declining effectiveness of widely used antibiotics and the emergence of new pathogenic microorganisms and/or the increasing resistance of existing ones (Skulberg, 2000). This means that it is becoming increasingly important to search for new sources of bioactive antimicrobial compounds. Cyanobacteria (blue-green algae) are considered to be a rich source of secondary metabolites that has yet to be fully explored (Ehrenreich et al., 2005). According to Skulberg (2000), the pharmaceutical effects and modes of action of cyanobacterial secondary metabolites are diverse (Thajuddin et al., 2005). These compounds include immunosuppressants and toxins, as well as antimicrobial and anti-tumor compounds (Ehrenreich et al., 2005; Barrios-Llerena et al., 2007). The importance of cyanobacteria as a potential drug resource is evidenced by the launch of the “Cyanomyces” project in Europe, which aims to generate novel therapeutic substances by combining genes from actinomycetes and cyanobacteria (http://www.cyanomyces.com) (Asthana et al., 2006). In addition, other microalgae have long been known to be potent sources of antimicrobial compounds (see, for example, Pratt et al., 1944 on Chlorella; Rossell et al., 1987 on brown algae). Among the chrysophytes, a chlorophyll-related bioactive compound called malhamensilipin A has been isolated from Poterioochromonas (Pereira et al., 2010). Porphyrin forms the core of chlorophyll and derivatives of this lipophilic compound include (Z)-24-propylidenecholesterol, which is proposed as a chemotaxonomic marker for some chrysophytes belonging to the class Pelagophyceae (Giner et al., 1998). Porphyrin derivatives have also been found in dinoflagellates and are thought to play a defensive role whereby their structural modifications make the sterols inedible to marine invertebrates, hindering predation and thereby enhancing the bloom-forming ability of dinoflagellates (Giner et al., 2003).
2In Egypt, local strains of microalgae are proving to represent quite promising potent sources of bioactive antimicrobial compounds (Issa, 1999; El-Sheekh et al., 2006), especially lipophilic microalgal extracts (El Semary et al., 2009; El Semary et al., 2010; El Semary, 2012). Our research follows on from these earlier studies by carrying out further screening of microalgae collected from the Helwan Governorate region of Egypt. In the present study, two microalgae were isolated, one belonging to the chrysophytes and the other to the cyanobacteria. First of all, we established the taxonomic identity of these microalgae. We then investigated the antimicrobial bioactivity of lipophilic extracts taken from both these microalgae and from another cyanobacterial strain, against many pathogenic microorganisms, some of them multidrug-resistant strains. The lipophilic extracts from the microalgal strains were then fractionated using column chromatography and the separated fractions were used in antimicrobial bioassay.
2. Materials and methods
2.1. Sampling and establishing microalgal cultures
3Water samples were collected in sterile samplers from the River Nile, in the Maadi area, and from a wastewater canal, in Helwan Governorate. Microscopic examination revealed the presence of cyanobacteria, diatoms, chrysophytes and some chlorophytes. Water samples were spun and spread on solid Oscillatoria medium (Feuillade, 1994) and ASN medium (50 ml medium: 0.5 gm agar, v: wt). Colonies were identified using light microscopy, and were then purified and used to establish monospecific cultures. Two microalgae were isolated: one a chrysophyte from the River Nile and one a cyanobacterium from Teraat El Khashaab. The two cultures were purified and identified morphologically using light microscopy.
2.2. Taxonomic identification of the two microalgae
4Genomic DNA was extracted using a modified method from Smoker et al. (1988). The small subunit rRNA gene was used as a taxonomic marker. For the cyanobacterium, the purified genomic DNA was used as a template for amplification of partial 16S rDNA using cyanobacterial highly-specific primers Cya395 and 781R1, 2 (Nübel et al., 1997). Almost the full length of 16S rDNA was then obtained using the Cya395 forward primer in combination with the 1492 reverse primer designed by Delong (1992). The taxonomic identity of the two microalgae was verified using Euk forward and Euk reverse primers (Delong, 1992). The 16S rRNA and 18S rRNA gene sequences were deposited in the GenBank database under the accession numbers JF442038 and JF442039.
5For the purpose of phylogenetic analyses, cyanobacterial 16S rRNA gene sequences were imported from GenBank and aligned in the Clustal W tool within the alignment function of the MEGA4 phylogenetic package. Similarly, the closely-related 18S rRNA genetic sequences of Ochromonas were used to reconstruct phylogenetic trees using different methods integrated into the MEGA4 software (Tamura et al., 2007). Bootstrap values from 500 resamplings were calculated for each set of data and all trees were rooted using the unicellular cyanobacterium Microcystis aeruginosa PCC7806 for the cyanobacterium and Scenedesmus communis for the Ochromonas. All resulting trees had similar topologies, indicating similar phylogenetic relationships. The bootstrapped-consensus maximum parsimony tree was chosen for presentation. The ME tree was searched using the Close-Neighbor-Interchange (CNI) algorithm (Nei et al., 2000). The Neighbor-Joining algorithm (Saitou et al., 1987) was used to generate the initial tree. Phylogenetic analyses were conducted using MEGA4 (Tamura et al., 2007).
2.3. Antimicrobial bioassay
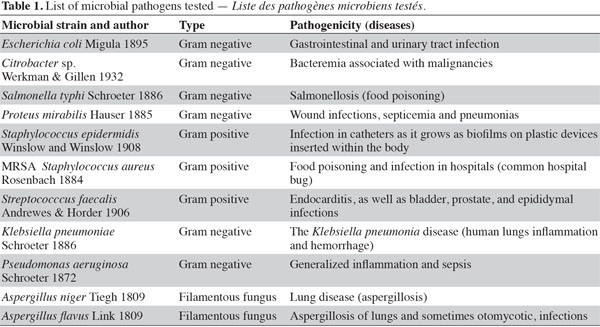
6Initial antibiotic susceptibility test. Pathogenic microbes (Table 1) were used as test microorganisms in antimicrobial tests (Doan et al., 2000). These test organisms were potent microbial pathogens collected from clinical samples and preserved as pure cultures. Their pathogenic effects are listed in table 1. The isolates were identified using traditional biochemical tests (Murray et al., 1995). The tested organisms were subcultured at 37 ºC and were maintained on nutrient agar media (Oxide, UK). Several antibiotics were tested in the initial susceptibility/resistance test. The bacterial isolates were tested for their resistance to antibiotics using a disk diffusion method on Mueller-Hinton agar (Oxide, UK) and were incubated overnight at 37 °C. The antibiotic disks (Oxoid, UK) used were ampicillin (10 μg), cefepime (30 μg), cefotaxime (30 μg), chloramphenicol (30 µg), doxycycline (30 µg), erythromycin (15 µg), levofloxacin (5 µg), rifampicin (30 µg), tobramycin (10 µg), ciprofloxacin (15 µg), amikacin (30 µg) amoxycillin/clavulinic acid (30 µg), oxacillin (30 µg), and ceftriaxone (30 µg). Zones of inhibition were determined in accordance with the National Committee for Clinical Laboratory Standards (NCCLS, 2000); isolates were categorized as either susceptible or resistant (Table 2). Results of the antibiotic susceptibility test are recorded in table 2.
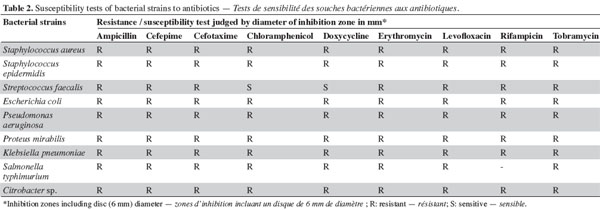
7Antimicrobial bioassay of lipophilic extracts. Microalgal biomass (1 g fresh wt.) was collected from both microalgae as well as from Aphanizomenon sp. and the samples were then lyophilized. The lyophilized cells were extracted twice with 100 ml chloroform: methanol (2:1, v: v) HPLC grade for two days, and then centrifuged (14,000 × g) for 30 min using a Hettich-cooling centrifuge (Germany). The supernatant was left to evaporate to dryness and was then re-dissolved in 3 ml chloroform: methanol (2:1, v: v) (modified from Doan et al., 2000). The sample was applied to a Silica gel G60 column (1.5 x 25 cm) prepared from the slurry from 30 g of precipitated Silica gel G60 (Merck, UK). The lipophilic fractions were collected using the solvent system shown in table 3.
8The lipophilic extracts were concentrated before being applied to 6 mm paper disks for testing. The paper was left to dry and for the solvent to evaporate before being used in the antimicrobial test. The sensitivity of these pathogenic strains to the extracted fractions was assessed using the slightly modified Kirby Bauer Disk Diffusion Susceptibility method (Bauer et al., 1966). The diameter of the inhibition zone was measured in triplicate and the average and standard deviation recorded (Table 4). Disks containing chloroform/methanol were left to evaporate and were then used as negative controls.

3. Results
3.1. Morphological identification
9Both microalgal strains isolated were cocooid cells and were either solitary or aggregated in clumps without a common sheath around them.
3.2. Small subunit rRNA gene molecular analysis and phylogenetic reconstruction
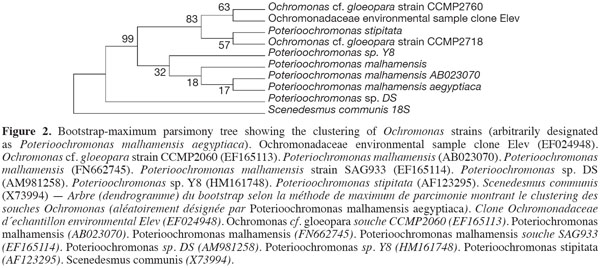
10The sequence retrieved was compared with other sequences deposited in GenBank using nucleotide BLAST search. The sequence showed a 97% similarity with the other sequences, with the best relative being Synechocystis salina LEGE 06099 (HQ832895). The other best relatives were Synechocystis Synechocystis salina LEGE 06155 (HQ832911), and Synechocystis PAK-12 (EF429297), both showing a 90% similarity. The phylogenetic analyses using different cyanobacterial representatives and employing different tree reconstruction methods all resulted in trees of similar topologies in which the isolate clustered with Synechocystis. They showed a distinct clade that separated away from the out-group taxa of Microcystis sp. and Lyngbya polychroa LP16 (FJ602745) (Figure 1). This clade was supported by a 91% bootstrap confidence value. The Synechocystis clade including the strain under study was further sub-clustered from the main sub-clade containing Synechocystis strains with a relatively large bootstrap value (67%). The chrysophyte strain showed a 97% similarity to Poterioochromonas malhamensis (AB023070), followed by Poterioochromonas malhamensis (FN662745), and Poterioochromonas sp. Y8 (HM161748). These strains formed a sub-cluster, supported by 99% bootstrap values, from the rest of the Ochromonas strains and the out-group taxon Scenedesmus communis (Figure 2).
3.3. Antimicrobial bioassay
11The preliminary antibiotic sensitivity tests showed the resistance of most of the pathogenic strains to the most commonly used antibiotics (Table 2). The only strain showing susceptibility to chloramphenicol and doxycycline was Streptococcus faecalis. The lipophilic fractions of Aphanizomenon sp. did not show any antibacterial bioactivity against Citrobacter sp., Escherichia coli, Proteus mirabilis, Staphylococcus epidermidis, MRSA Staphylococcus aureus, Streptococccus faecalis, Klebsiella pneumonia or Pseudomonas aeruginosa. Fraction V showed antifungal bioactivity against both Aspergillus niger and Aspergillus flavus. Fractions IV and VI showed little antifungal bioactivity against Aspergillus flavus (Table 3). The lipophilic fractions of Synechocystis salina were not effective against Proteus mirabilis, Staphylococcus epidermis, MRSA Staphylococcus aureus, Streptococccus faecalis, Klebsiella pneumonia, Pseudomonas aeruginosa or the pathogenic fungus Aspergillus flavus. The only fraction that was effective against Aspergillus niger was fraction V. Two fractions were effective against Citrobacter sp. (fractions III and IV). These fractions, together with fraction II, were effective against Salmonella typhi (Table 3). The lipophilic fractions of Poterioochromonas malhamensis were not effective against any fungal strain or against most of the bacterial strains, except for Citrobacter sp. and Salmonella typhi. Fractions III.1 and III.2 were highly effective against these last two bacterial strains (Table 4).
4. Discussion
12The identity of the local microalgal strains (Synechocystis salina, a prokaryotic coccoid cyanobacterium and Poterioochromonas malhamensis, a eukaryotic microalga belonging to the chrysophytes) was confirmed by traditional phenotypic characterization and modern molecular and phenotypic analyses. The high percentage of sequence identities of both local strains and their co-clustering among similar isolates of known identities further confirmed their taxonomic designations. Poterioochromonas malhamensis is known for its mixed mode of nutrition whereby phagotrophy is performed in the form of bacteriovory along with phototrophy. This mixed mode of nutrition is very important in aquatic systems as it ensures the high survival rate of this microalga even under stress conditions. It also means that high death rates of bacteria can be attributed to the activity of this microalga (Sanders et al., 1990). Supernatants from microalgal cultures of Poterioochromonas malhamensis have been shown to exhibit antibiotic action that may be mostly attributed to lipophilic substances released from this microalga (Blom et al., 2010). In addition, extracts from Poterioochromonas malhamensis have shown a broad-spectrum antibacterial effect (Blom et al., 2010). Moreover, the bioactivity of one compound, called malhamensilipin A, isolated from this microalga has been reported (Pereira et al., 2010). Malhamensilipin A acts as an antiviral and antimicrobial agent as well as an inhibitor of the enzyme tyrosine kinase, which also indicates its possible activity as an anticancer agent (Chen et al., 1994). The microalga Poterioochromonas malhamensis itself is reported to possess several chlorine-substituted bioactive compounds exhibiting antimicrobial activity (Chen et al., 1994). This may explain the antimicrobial bioactivity of extracts from this microalga against multidrug-resistant strains in the present study.
13The second microalgal strain under study, Synechocystis salina, was isolated from a highly-polluted wastewater canal into which industrial, agricultural and domestic wastes are known to be released. This has resulted in water of a brackish nature (conductivity value of 8,091 μ·Scm-1) (Hamed, 2008) and the prevalence of stressful conditions that select for microalgae with unique metabolic features, enabling them to survive and thrive (El Semary, 2012). Interestingly, in his study of Egyptian water habitats, including water bodies in the Helwan area, Hamed (2008) found that Synechocystis salina thrived within the brackish-saline-hypersaline range of conductivity in a wide range of habitats. This indicates the possibility that the metabolic adaptability of this cyanobacterium is mediated through the possession of unique metabolites.
14Our results also showed the extreme resistance of most of the isolates tested to the most commonly used antibiotics. The emergence of microbial multidrug-resistance to antibiotics is attributed to many complex factors, such as mutations, genetic recombinations and geographical segregation (Hujer et al., 2006). Microbial multidrug resistance represents a widespread problem nowadays, especially where antibiotics have been intensively used (Piddock, 2002). Pathogenic bacteria develop resistance by acquiring genes that encode proteins to protect them from the effects of the antibiotic. In some cases, the genes arise through mutation, but in others they are acquired from other bacteria that are already resistant to antibiotics. These genes are often found on plasmids, which spread easily from one bacterium to another (Fluit et al., 2001; Bhakta et al., 2003).
15In the present study, although extracts were relatively effective against some pathogens, no single extract had a broad-spectrum effect against all the pathogenic strains studied. Nevertheless, a few extracts showed some cross antimicrobial bioactivity against different microorganisms, e.g. fraction IV of Aphanizomenon sp. (Table 4). El Semary (2009) conducted phytochemical screening for Aphanizomenon sp., showing that the bacterium possessed alkaloids and flavonoids, and that it had a relatively high lipid content as well as a high level of iron. Interestingly, in the same study, molecular screening for the presence of genetic loci of secondary metabolites was positive for Aphanizomenon sp., indicating its potentiality as a producer of bioactive compounds and as a possible antibiotic resource. This hypothesis was then confirmed by the current investigation.
16In conclusion, the extracts from the microalgae studied here showed considerable antimicrobial bioactivity against several multidrug-resistant isolates. Screening programs need to be extended further in order to discover more bioactive compounds from local microalgal strains in Egypt.
17Acknowledgements
18The authors would like to express their deepest gratitude to Dr Seraya Maouche from Universitätsklinikum Schleswig-Holstein and Dr Nahla El Sherif, Department of Botany, Faculty of Science, Ain Shams University, Egypt for the great help with French translation and Dr David Adams, University of Leeds for proofreading the manuscript.
Bibliographie
Asthana R.K. et al., 2006. Identification of an antimicrobial entity from the cyanobacterium Fischerella sp. isolated from bark of Azadirachta indica (neem) tree. J. Appl. Phycol., 18, 33-39.
Barrios-Llerena M.E., Burja A.M. & Wright P.C., 2007. Genetic analysis of polyketide synthase and peptide synthetase genes in cyanobacteria as a mining tool for secondary metabolites. J. Ind. Microbiol. Biotechnol., 34, 443-456.
Bauer A.W., Kirby W.M., Sherris J.C. & Turck M., 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol., 45, 493-496.
Bhakta M., Arora S. & Bal M., 2003. Interspecies transfer of a chloramphenicol-resistance plasmid of staphylococcal origin. Indian J. Med. Res., 117, 146-151.
Blom J.F. & Pernthaler J., 2010. Antibiotic effects of three strains of chrysophytes (Ochromonas, Poterioochromonas) on freshwater bacterial isolates. FEMS Microbiol. Ecol., 71(2), 281-290.
Borowitzka M.A., 1995. Microalgae as sources of pharmaceuticals and other biologically active compounds. J. Appl. Phycol., 7, 3-15.
Chen J.L. et al., 1994. Structure of malhamensilipin A, an inhibitor of protein tyrosine kinase, from the cultured chrysophyte Poterioochromonas malhamensis. J. Nat. Prod., 57(4), 524-527.
Delong E.F., 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U.S.A., 98, 5685-5689.
Doan N.T., Rickards R.W., Rothschild J.M. & Smith G.D., 2000. Allelopathic actions of the alkaloid 12-epi-Hapalindole E isonitrile and calothrixin A from cyanobacteria of the genera Fischerella and Calothrix. J. Appl. Phycol., 12, 409-416.
Ehrenreich I.M., Waterbury J.B. & Webb E.A., 2005. Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes. Appl. Environ. Microbiol., 71(1), 7401-7413.
El Semary N.A., 2009. Investigating the potential for biodiesel and bioactive compounds production from some cyanobacteria. Egypt. J. Phycol., 10, 85-97.
El Semary N.A., 2012. The characterisation of bioactive compounds from an Egyptian Leptolyngbya sp. strain. Ann. Microbiol., 62, 55-59.
El Semary N.A., Ghazy S.M. & Abd El Naby M., 2009. Investigating the taxonomy and bioactivity of an Egyptian Chlorococcum isolate. Aust. J. Basic Appl. Sci., 3(3), 1540-1551.
El Semary N.A. & Abd El Naby M., 2010. Characterization of a Synechocystis sp. from Egypt with the potential of bioactive compounds production. World J. Microbiol. Biotechnol., 26(6), 1125-1133.
El-Sheekh M.M., Osman M.E.H., Dyabb M.E. & Amer M.S., 2006. Production and characterization of antimicrobial active substance from the cyanobacterium Nostoc muscorum. Environ. Toxicol. Pharmacol., 21, 42-50.
Feuillade J., 1994. The cyanobacterium (blue-green alga) Oscillatoria rubescence D.C. Arch. Hydrobiology, 42, 77-93.
Fluit A.D.C., Maarten R.V. & Schmitz F.J., 2001. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev., 14, 836-871.
Giner J.-L. & Boyer G., 1998. Sterols of the brown tide alga Aureococcus anophagefferens. Phytochemistry, 48(3), 475-477.
Giner J.-L., Faraldos J.A. & Boyer G., 2003. Novel sterols of the toxic Dinoflagellate Karenia brevis (Dinophyceae): a defensive function for unusual marine sterols. J. Phycol., 39(2), 315-319.
Hamed A.F., 2008. Biodiversity and distribution of blue-green algae/cyanobacteria and diatoms in some of the Egyptian water habitats in relation to conductivity. Aust. J. Basic Appl. Sci., 2(1), 1-21.
Hujer K.M. et al., 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother., 50(12), 4114-4123.
Issa A.A., 1999. Antibiotic production by the cyanobacteria Oscillatoria augustissima and Calothrix parietina. Environ. Toxicol. Pharmacol., 8, 33-37.
Mundt S., Kreitlow S. & Jansen R., 2003. Fatty acids with antibacterial activity from the cyanobacterium Oscillatoria redekei HUB 051. J. Appl. Phycol., 15, 263-267.
Murray P.R.E. et al., 1995. Manual of clinical microbiology. 6th ed. Washington, DC, USA: American Society for Microbiology Press.
NCCLS (National Committee for Clinical Laboratory Standards), 2000. Performance standard for antimicrobial disk susceptibility tests. 7th ed. Villanova, PA, USA: NCCLS, Approved Standard: M2-A7.
Nei M. & Kumar S., 2000. Molecular evolution and phylogenetics. New York, USA: Oxford University Press, http://www.kumarlab.net/publications, (05/01/2013).
Nübel U., Garcia-Pichel F. & Muyzer G., 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol., 63(8), 3327-3332.
Pereira A.R. et al., 2010. Structure revision and absolute configuration of malhamensilipin A from the freshwater chrysophyte Poterioochromonas malhamensis. J. Nat. Prod., 73(2), 279-283.
Piddock L.J.V., 2002. Fluoroquinolone resistance in Salmonella serovars isolated from humans and food animals. FEMS Microbiol. Rev., 26, 3-16.
Pratt R.H. et al., 1944. Chlorellin, an antibacterial substance from Chlorella. Science, 99, 351-352.
Rossell K.G. & Srivastava L.M., 1987. Fatty acids as antimicrobial substances in brown algae. Hydrobiologia, 151/152, 471-475.
Saitou N. & Nei M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406-425.
Sanders R.W., Porter K.G. & Caron D.A., 1990. Relationship between phagotrophy and phototrophy in the mixotrophic chrysophyte Potrioochromonas malhamensis. Microb. Ecol., 19, 97-109.
Skulberg O.M., 2000. Microalgae as a source of bioactive molecules – experience from cyanophyte research. J. Appl. Phycol., 12, 341-348.
Smoker J.A. & Baruum S.R., 1988. Rapid small-scale DNA isolation from filamentous cyanobacteria. FEMS Microbiol. Lett., 56, 119-222.
Tamura K., Dudley J., Nei M. & Kumar S., 2007. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol., 24, 1596-1599.
Thajuddin N. & Subramanian G., 2005. Cyanobacterial biodiversity and potential applications in biotechnology. Curr. Sci., 89(1), 47-57.
Whitton B.A. & Potts M., 2000. Freshwater blooms. In: Whitton B.A. & Potts M., eds. The ecology of cyanobacteria: their diversity in time and space. Dordrecht, The Netherlands: Kluwer Academic Publishers.
