- Home
- Volume 19 (2015)
- numéro 3
- Efficient genetic transformation of chicory without selection marker
View(s): 1470 (18 ULiège)
Download(s): 99 (2 ULiège)
Efficient genetic transformation of chicory without selection marker

Editor's Notes
Received on August 9, 2014; accepted on April 13, 2015
Résumé
Transformation génétique efficace de la chicorée sans pression de sélection
Description de l'objet. La présente étude explique une méthode de transformation au moyen d'Agrobacterium pour la chicorée basée sur la régénération des pousses sur un milieu ne contenant pas de composés sélectifs (une approche non sélective).
Objectifs. Le but de cette expérience était de générer des plantes transgéniques de chicorée sur un milieu d'induction de pousses sans composés sélectifs et d’étudier la transmission du transgène intégré.
Méthode. Des explants de feuilles ont été collectés à partir de plants cultivés in vitro du cultivar ‘ Melci ’. Les explants ont été inoculés avec Agrobacterium hébergeant le plasmide pTJK136. L’induction de cals et la régénération des pousses ont été effectuées sur le milieu ne contenant pas d'antibiotiques. Trois expériences de transformation séparées ont été effectuées. La présence du transgène dans des plantes régénérées (T0 et T1) a été examiné par PCR et RT-PCR. En outre, la fréquence de régénération et l'efficacité de transformation sont calculées séparément pour chaque expérience.
Résultats. Les résultats d'analyse indiquent que des nombres importants de pousses régénérées récupérés sur un milieu ne contenant aucun agent de sélection sont des transformants potentiels. Le criblage des plantes régénérées à partir des expériences en triplicat a révélé une remarquable efficacité de transformation (jusqu'à 9 %). La PCR après transcription inverse (RT-PCR) a confirmé l'expression correcte du transgène dans les plantes transformées et donc une intégration stable dans le génome. De plus, ce dernier point est corroboré par le fait que le transgène est transmis avec succès à la prochaine génération (T1).
Conclusions. Les résultats ont confirmé la possibilité de générer des plantes génétiquement modifiées sans recours à un marqueur de sélection lors de la transformation.
Abstract
Description of the subject. The present study explains an Agrobacterium-mediated transformation method for chicory based on shoot regeneration medium containing no selective compounds (a non selection approach).
Objectives. The aim of this experiment was to generate transgenic chicory plants on a shoot induction medium without selective compounds and study the inheritance of the integrated transgene.
Method. Leaf explants were collected from in vitro grown seedlings of cultivar ‘Melci’. Explants were inoculated with Agrobacterium harboring the pTJK136 plasmid. Callus induction and shoot regeneration were performed on the medium containing no antibiotics. Three separate transformation experiments were carried out. The presence of the transgene in regenerated plants (T0 and T1) was examined by PCR and RT-PCR. Furthermore, the regeneration frequency and transformation efficiency were separately calculated for each experiment.
Results. Analytical results indicate that significant numbers of regenerated shoots recovered on a medium containing no selection agent are putative transformants. Screening of the regenerated plants from triplicate experiments revealed a remarkable transformation efficiency (up to 9%). Reverse transcription PCR (RT-PCR) analysis confirmed the correct expression of the transgene in the transformed plants and thus a stable integration in the genome. Additionally, to corroborate this last point, the transgene is successfully transmitted to the next generation (T1).
Conclusions. Results confirmed the possibility of generating marker-free genetically modified plants in the form of a non-selection transformation system.
Table of content
1. Introduction
1The tuberous root of chicory plants Cichorium intybus contains a number of medicinally important compounds such as inulin, sesquiterpene lactones, coumarins, flavonoids and vitamins (Nandagopal et al., 2007). Chicory root is mostly cultivated for the production of inulin (De Bruyn et al., 1992). Inulin, an important storage carbohydrate, is used in the food industry because of its large number of health promoting functions (Kaur et al., 2002). Therefore, improvement and development of new cultivars with higher levels of inulin or other nutritional components are necessary. Since genetic improvement of crops by selection methods may take several generations, transgenic technology has become a valuable tool due to its quick methodology. Genetic engineering of plants mostly involves the transfer of genes to the plant genome followed by regeneration leading to the production of transgenic plants (Tuteja et al., 2012). However, the plant transformation methods by Agrobacterium, microinjection, particle gun, or protoplast transformation are generally considered as relatively inefficient (Rakoczy-Trojanowska, 2002). Therefore, a selectable marker gene, such as neomycin phosphotransferase II (nptII) hygromycin phosphotransferase (hpt) and phosphinothricin acetyltransferase (bar), is generally co-introduced with the gene of interest to identify the transformed plant cell and eventually the regenerated plant (Lee et al., 2008). However, existence of such marker selection genes in the genetically modified crops may render the crop less acceptable to the consumers (Miki et al., 2004; Bala et al., 2013). Although there are a number of methods available for the removal of selectable marker genes from transgenic plants but the direct generation of marker-free transgenic plants is a better way to address public concern over the safety of genetically engineered (GE) crops. To consider this important issue, development of an efficient marker-free transgenic system for chicory is now essential. Clean vector technology using a non-selection approach intends to produce GE plants with only the gene-of-interest as newly introduced functional gene without any superfluous genes (Krens et al., 2004). Thus, it is useful to develop transformation without introducing a selectable marker gene and screening only for the gene of interest. In this study, genetic modification with Agrobacterium harboring the gene of interest followed by regeneration of shoots under no selection method aimed at producing transgenic chicory plants.
2. Materials and methods
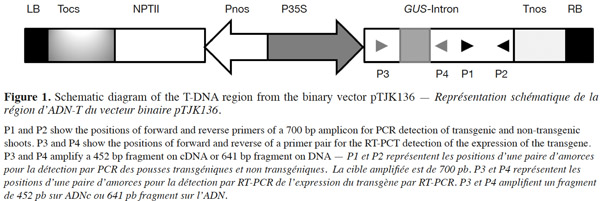
2.1. Plant material and Agrobacterium strain
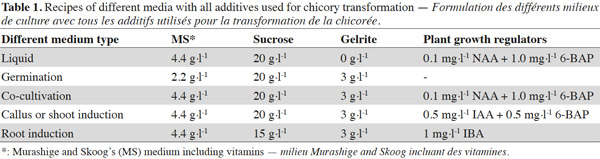
2The seeds of C. intybus cultivar ‘Melci’ were surface-sterilized. Aseptic seeds were cultured on germination medium (Table 1) at 26 ± 1 ºC under 16 h photoperiod and a 1,000-1,200 lux light intensity to germinate and then first leaves were used to prepare explants. Agrobacterium tumefaciens strain C58C1 RifR (pGV2260) (Deblaere et al., 1987), harboring the shuttle vector pTJK136 was used in this study. pTJK136 is carrying the intron-containing β-glucuronidase (uidA or GUS) gene under the control of the CaMV35S promoter and 3' nos terminator (Kapila et al., 1997; Figure 1) and in this study it will be considered as the model gene of a new trait to be introduced in chicory. The Agrobacterium was inoculated into a YEB medium (Vervliet et al., 1975) containing 50 mg·l-1 rifampicin, 100 mg·l-1 carbenicillin and 100 mg·l-1 spectinomycin. The inoculated YEB medium was incubated at 28 °C with shaking (200 cycles·min-1) for 24 h. The culture was transferred to a sterile 50 ml tube, and then centrifuged at room temperature for 10 min at 3,000 g. After removing the supernatant, the pellet was gently resuspended in 100 ml of sterile liquid medium (Table 1) supplemented with 20 mg·l-1 acetosyringone for inoculation of explants. The cell suspension to be reached as an OD600 = 1.
2.2. Plant regeneration and transformation
3Leaf explants were prepared from 21-day-old in vitro seedlings. After wounding on the abaxial surfaces of explants, they were immersed for 30 min in the liquid medium supplemented with the suspension of Agrobacterium strain harboring the pTJK136 plasmid. Compositions of all the media were based on Murashige and Skoog (1962) formulation (Table 1). Inoculated explants were blotted dry on sterile filter paper and transferred to co-cultivation medium (Table 1) containing no antibiotic. Eight to ten leaf pieces were placed on co-cultivation medium in a 9 cm Petri dish. Co-cultivated explants were incubated for 3 days in growth chamber under 16/8 h (light/dark) photoperiod with white fluorescent lights giving a photon flux density of 150–200 µM·m-2·s-1 at a temperature of 24 °C ± 2. After co-cultivation, the explants were washed 3 times in sterilized liquid medium (Table 1) supplemented with 500 mg·l-1 cefotaxime to counterselect Agrobacterium cells. The explants were then dried on sterile filter paper and transferred onto callus/shoot induction medium (Table 1) supplemented with 500 mg·l-1 cefotaxime. The selective agent kanamycin was not included in the callus/shoot induction medium. After callus induction, the explants were sub-cultured every two weeks on fresh callus/shoot induction medium containing 500 mg·l-1 cefotaxime until shoots appeared. Regenerated shoots were excised from the explants and individually transferred into 175 ml glass jars containing root induction medium (Table 1). The shoots with suitable root system were gently removed from root induction medium and washed in liquid medium to clean the gelling pieces. The well developed plantlets were then transferred into 10-15 cm pots containing sterilized-soil. Pots were covered with a transparent plastic and placed in the growth chamber to acclimatize. After 2-3 days, plastic cover was opened from the top and after 1 week, when the plants were strong enough, the transparent plastic cover was completely removed. Three separate transformation experiments were carried out, in the first experiment 100 explants, in the second 150 and in the third 120 explants were inoculated. The regeneration frequency and transformation efficiency were calculated for each experiment by the formulas given below (Zaidi et al., 2006):

4The transgenic nature and transmission of the transgene to the next generation (T1) was also studied. A few roots of original transgenic (T0) plants were firstly stored for three months at 0-4 °C and 80-90% humidity. The roots were replanted in the spring for seed production. When plants produced flowers, they were fully covered with bags. The T0 plants were next self-crossed by hand-pollination. After the seeds were developed, they were harvested for further assessment.
2.3. DNA/RNA extraction and PCR analysis
5Total genomic DNA was isolated from leaf tissue of regenerated and untransformed control plants using a modified CTAB extraction procedure (Doyle, 1991). Total RNA was also extracted from leaves of regenerated and untransformed control plants following the guanidiniumthiocyanate-phenol-chloroform isolation method described previously (Chomczynski et al., 2006). To eliminate possible genomic DNA contamination, total RNA was treated with DNase (Vivantis) according to the manufacturer’s instructions. Five hundred ng of uncontaminated total RNA were used to make first-strand cDNA with the Viva 2-steps RT-PCR Kit (Vivantis) using oligo-dT primers. The quantity and quality of total RNA or DNA were determined using a Nanodrop ND1000 spectophotometer. A specific primer pair (forward: 5’-TGCAACTGGACAAGGCACTA-3’; reverse: 5’-ATCGCTGATGGTATCGGTGT-3’) was designed to amplify a 700 bp fragment on uidA gene (Figure 1). A specific pair of primer (forward: 5’-GCCAGCGTATCGTGCTGCGT-3’; reverse: 5’-GCTAGTGCCTTGTCCAGTTG-3’) was also designed to amplify a 452 bp fragment in case of intron splicing on cDNA or to amplify a 641 bp fragment on genomic DNA or Agrobacterium contamination (Figure 1). PCR amplifications were performed by designed specific primers on either DNA or cDNA templates. Amplification products were then separated by electrophoresis on 1.2% agarose gel and stained with ethidium bromide and visualized under UV trans-illuminator. PCR reactions were carried out essentially as follows: 100 ng of DNA or cDNA, 15 pmol of each primer, 200 µM dNTPs mix, 5 µl PCR buffer 10x, 1 unit Taq DNA polymerase and autoclaved Millipore water in final volume of 50µl in a Bio-Rad thermocycler. PCR conditions were 2 min at 95 °C; followed by 35 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at specific temperature for each primer pair and extension according to 60 s for 1 kb at 72 °C and ending with 7 min at 72 °C for final extension.
3. Results
3.1. Plant regeneration
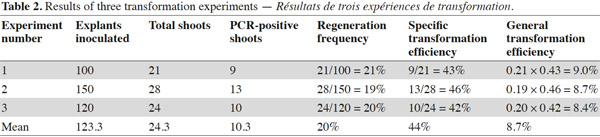
6Shoot regeneration has been achieved in shoot induction medium containing no antibiotic on leaf explant-derived calli inoculated with Agrobacterium. The regenerated shoots were cut and transferred to root induction medium. Roots appeared after 2-3 weeks from cutting surfaces. All rooted plantlets were transferred into 10-15 cm pots covered with a transparent plastic and located in the growth chamber for adaptation. Recovered plants were then transferred to greenhouse for further growth and analysis. The results of three experiments are summarized as shown in table 2. After co-cultivation of explants with Agrobacterium and regeneration, the following frequencies of regenerated shoots were obtained (Table 2): 21/100 (21.0%) in experiment 1, 28/150 (19.0%) in experiment 2 and 24/120 (20.0%) in experiment 3 or in other words a global result of 73 regenerated shoots from 370 explants (20%). Moreover, specific transformation efficiency and global transformation efficiency were separately calculated for each experiment (Table 2). The resulting data from the three experiments produce replicable evidence indicating the efficiencies are valid. Additionally, from few T0 plants self-seeds were harvested and analyzed for patterns of transgene integration.
3.2. Molecular characterization of regenerated plants and screening of their progeny
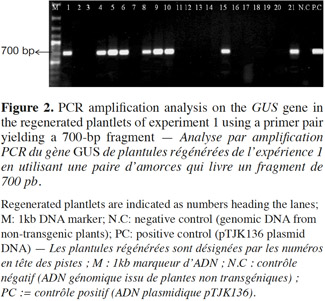
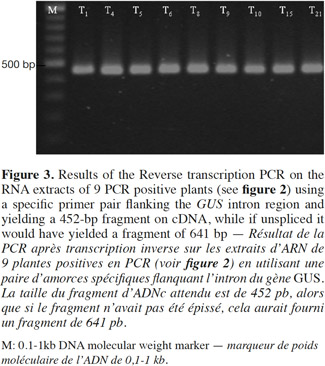
7Presence of the transgene in the regenerated plants was first examined by PCR analysis on genomic DNA using specific primers to amplify the expected 700 bp fragment on uidA (Figure 1). The banding pattern of PCR amplified products from regenerated shoots was obtained. The expected band was amplified in the plantlets defined as transgenic and no GUS band was amplified in the non-transformed plantlets (Figure 2). Additionally RT-PCR was performed to amplify a 452 bp in case of intron splicing on cDNA or to amplify 641 bp on Agrobacterium contamination (Figure 1). Since the GUS-intron gene was transferred, amplification of the 452 bp fragment by RT-PCR analysis confirms expression of the transgene in the transformants (Figure 3). The transformation efficiencies were calculated using the percentage of the regenerated plants (T0 plants) that were positive at the PCR detection test. Transformation efficiencies obtained in the three experiments were 9, 8.7 and 8.4% respectively (Table 2). Therefore the three experiments verified statistically the result. The progenies (T1) of original transgenic plants (T0) were also assessed to study inheritance of the new trait. A number of T1 progenies were subjected to RT-PCR to detect the presence or absence of the transgene (Figure 4). RT-PCR assay showed the expression of uidA by amplification of the expected 452 bp indicating inheritance and segregation of transgene in the T1 populations at a ratio which can be in line with an expected 3:1 Mendelian segregation ratio. Due to the limited number of offsprings analyzed (Figure 4), it is difficult to state if the slight overrepresentation of segregants with the transgenic trait is significant or not. Would it however be so then multiple insertion could account for it.

4. Discussion
8Transgenic crops containing no marker gene are an essential requirement for successful and rapid commercialization. Possibility of uncontrolled spreading in natural plant population via cross-pollination or microorganisms by the way of horizontal gene transfer is a limiting factor as well (Tuteja et al., 2012). On account of these matters, a number of approaches, such as site-specific recombinase-mediated excision systems (Dale et al., 1991; Gleave et al., 1999; Zuo et al., 2001), co-transformation and segregation (Depicker et al., 1985; McKnight et al., 1987; Komari et al., 1996; Daley et al., 1998), transposon-mediated reposition (Goldsbrough et al., 1993), intrachromosomal homologous recombination (Puchta, 2000; Zubko et al., 2000), and generation of selectable marker-free transgenic plants under no selection (Dominguez et al., 2002; De Vetten et al., 2003) have been developed to produce marker-free transgenic plants. However, the natural method to obtain marker-free plants is transformation without any selectable marker gene (Chong-Perez et al., 2013). Generation of marker-free transgenic plants under no selection is less expensive and time consuming, even though more inefficient in comparison with other methods (Li et al., 2009). Additionally, in co-transformation and segregation method, the crop should be amenable to subsequent sexual crossing for segregating selection gene from target gene (Krens et al., 2003). Besides, cells with insertion events in silent regions like telomeres possibly do not survive when selection is applied (Chong-Perez et al., 2013).
9Transformation without the use of a selectable marker method, markerless method, from the initial stages on was thought to be impossible due to no or very low regeneration frequency of transformed shoots (De Buck et al., 1998). However, there are now a number of reports of the successful generation of marker free transgenic plants (Dominguez et al., 2002; De Vetten et al., 2003; Permingeat et al., 2003; Doshi et al., 2007; Jia et al., 2007; Xingguo et al., 2008; Li et al., 2009; Bhatnagar et al., 2010; Xin et al., 2012). Considering the public concerns regarding transgenic plants harboring antibiotic or herbicide resistant genes and also to make available a transformation method under utilization of no antibiotic or herbicide selective agents, the production of transgenic chicory under no selection approach was intended. The results here verified that it is possible to obtain transgenic chicory plants using an Agrobacterium-mediated transformation method that excludes the use of any selective agent. Besides, further analyses of the T0 progenies confirmed the transmission of the transgene to the next generation. The efficiency of regenerated transgenic chicory plants under no selection in comparison with other plants such as potato (De Vetten et al., 2003), tobacco (Li et al., 2009), apple (Malnoy et al., 2010) and tomato (Xin et al., 2012), seems rather remarkable. This could be elucidated by the very efficient regeneration or transformation system. Maroufi (2010) also used no selection system (no phosphinothricin as the selective agent) and obtained transgenic chicory plants (cultivar ‘Hera’) in the case a bar gene as a selectable marker was carried on the constructed T-DNA binary vectors. Data obtained from this study might provide evidence concerning producing transgenic plants in the absence of selectable marker genes.
10The frequencies of PCR positive transformants in three experiments indicate that the use of a selectable marker gene in the Agrobacterium-mediated transformation process for chicory can be avoided and therefore this method can be a practical method. Here, for the first time the production of chicory transgenic plants under no selection with rather high general transformation efficiency (= 8.7%) is reported. This could be interesting in comparison with the regeneration in the presence of antibiotic, which resulted an efficiency of 16% of transformed chicory plants (Maroufi et al., 2012). Selective agents such as phosphinothricin or kanamycin are limiting the ability of transgenic cells to proliferate and differentiate into transgenic plants, thus a non-selection approach limits the negative impacts on regenerated transgenic plants. Moreover, it is a single step process without involving the genetic segregation that could reduce the cost and time to produce marker free transgenic plants. Besides, due to the limiting numbers of available selectable marker genes, this achievement will be valuable for the stacking of transgenes in future (Tuteja et al., 2012). Since a few of constitutive promoters are commonly used to drive marker genes, therefore introduction of the same sequences could trigger gene-silencing mechanisms and may result in reduced expression of one or more transgenes (Puchta, 2003). Consequently, availability of a marker-free transformation system can provide better option for multiple transgenes pyramiding (Bhatnagar et al., 2010).
11In conclusion the markerless technique has several advantages over other techniques; it is relatively inexpensive, simple, quick, and does not require genetic segregation (De Vetten et al., 2003). Moreover, once the transgene or its expression is identified at the progenies, the marker-free transgenic plants are confirmed. Thus it is not required to do additional transformation or crossing (Li et al., 2009). This experiment was practical because a significant number of regenerated plants in the absence of selective agents are transgenic. The results were highly reproducible according to three separate experiments and do not need any more optimization of regeneration protocol. The transgene was able to be expressed in T0 and T1 generations under no selection showing by RT-PCR in a similar fashion as those recovered under selection. Finally, such transformation system has the potential for routine use in production of selectable marker-free transgenic chicory plants for research or commercial projects. This system which provided a quick and low-cost way of generating marker-free transgenic chicory is possibly of interest also in other plants.
12List of abbreviations
136-BAP: 6-benzylaminopurine
14CaMV: cauliflower mosaic virus
15CTAB: cetyltrimethyl ammonium bromide
16GE: genetically engineered
17IAA: indole-3 acetic acid
18IBA: indole-3 butyric acid
19NAA: 1-naphthaleneacetic acid
20nos terminator: nopaline synthase terminator
21OD: optical density
22RT-PCR: reverse transcription polymerase chain reaction
23YEB: yeast-extract broth
24Acknowledgements
25This work was supported by University of Kurdistan, Sanandaj, Iran. I would like to thank Plant System Biology, Ghent, Belgium for providing Agrobacterium and Joost Bart (ILVO, Ghent, Belgium) for providing chicory seeds.
Bibliographie
Bala A. et al., 2013. Development of selectable marker free, insect resistant, transgenic mustard (Brassica juncea) plants using Cre/lox mediated recombination. BMC Biotechnol., 13, 88.
Bhatnagar M. et al., 2010. An efficient method for the production of marker-free transgenic plants of peanut (Arachis hypogaea L.). Plant Cell Rep., 29, 495-502.
Chomczynski P. & Sacchi N., 2006. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat. Protoc., 1, 581-585.
Chong-Perez B. & Angenon G., 2013. Strategies for Generating Marker-Free Transgenic Plants, http://www.intechopen.com/books/genetic-engineering/strategies-for-generating-marker-free-transgenic-plants, (22/5/2013).
Dale E.C. & David W.O., 1991. Gene transfer with subsequent removal of the selection gene from the host genome. Proc. Natl. Acad. Sci. U.S.A., 88, 10558-10562.
Daley M., Knauf V.C., Summerfelt K.R. & Turner J.C., 1998. Co-transformation with one Agrobacterium tumefaciens strain containing two binary plasmids as a method for producing marker-free transgenic plants. Plant Cell Rep., 17, 489-496.
De Bruyn A., Alvarez A.P., Sandra P. & De Leenheer L., 1992. Isolation and identification of O-ß-D-fructofuranosyl-(2, 1)-O-ß-D- fructofuranosyl-(2, 1)-D-fructose, a product of the enzymic hydrolysis of the inulin from Cichorium intybus. Carbohydr. Res., 235, 303-308.
De Buck S., Jacobs A., Van Montagu M. & Depicker A., 1998. Agrobacterium tumefaciens transformation and co-transformation frequencies of Arabidopsis thaliana root explants and tobacco protoplasts. Mol. Plant-Microbe Interact., 11, 449-457.
De Vetten N. et al., 2003. A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nat. Biotechnol., 21, 439-442.
Deblaere R. et al., 1987. Vectors for cloning in plant cells. In: Wu R. & Grossman L., eds. Recombinant DNA, part D. Methods in enzymology. New York, USA: Academic Press, 277-292.
Depicker A. et al., 1985. Frequencies of simultaneous transformation with different T-DNAs and their relevance to the Agrobacterium plant cell interaction. Mol. Gen. Genet., 201, 477-484.
Dominguez A. et al., 2002. Regeneration of transgenic citrus plants under non selective conditions results in high-frequency recovery of plants with silenced transgenes. Mol. Genet. Genomics, 267, 544-556.
Doshi K.M., Eudes F., Laroche A. & Gaudet D., 2007. Anthocyanin expression in marker-free transgenic wheat and triticale embryos. In Vitro Cell Dev. Biol. Plant, 43, 429-435.
Doyle J.J., 1991. DNA protocols for plants-CTAB total DNA isolation. In: Hewitt G.M. & Johnston A., eds. Molecular techniques in taxonomy. Berlin: Springer, 283-293.
Gleave A.P., Mitra D.S., Mudge S.R. & Morris B.A.M., 1999. Selectable marker-free transgenic plants without sexual crossing: transient expression of cre recombinase and use of a conditional lethal dominant gene. Plant Mol. Biol., 40, 223-235.
Goldsbrough A.P., Lastrella C.N. & Yoder J.I., 1993. Transposition mediated re-positioning and subsequent elimination of marker genes from transgenic tomato. Biotechnology, 11, 1286-1292.
Jia H., Liao M., Verbelen J.P. & Vissenberg K., 2007. Direct creation of marker-free tobacco plants from agro infiltrated leaf discs. Plant Cell Rep., 26, 1961-1965.
Kapila J., De Rycke R., Van Montagu M. & Angenon G., 1997. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci., 122, 101-108.
Kaur N. & Gupta A.K., 2002. Applications of inulin and oligofructose in health and nutrition. J. Biosci., 27, 703-714.
Komari T. et al., 1996. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J., 10, 165-174.
Krens F.A. et al., 2003. Clean vector technology for marker-free transgenic crops. In: Tuberosa R., Phillips R.L. & Gale M., eds. Proceedings of the International Congress, In the wake of the double helix: from the green revolution to the gene revolution, 27-31 May, 2003, University of Bologna, Bologna, Italy, 509-516.
Krens F.A. et al., 2004. Clean vector technology for marker-free transgenic ornamentals. Acta Hortic., 651, 101-105.
Lee L.Y. & Gelvin S.B., 2008. T-DNA binary vectors and systems. Plant Physiol., 146, 325-332.
Li B., Xie C. & Qiu H., 2009. Production of selectable marker-free transgenic tobacco plants using a non-selection approach: chimerism or escape, transgene inheritance, and efficiency. Plant Cell Rep., 28, 373-386.
Malnoy M. et al., 2010. Genetic transformation of apple (Malus x domestica) without use of a selectable marker gene. Tree Genet. Genomes, 6, 423-433.
Maroufi A., 2010. Biotechnological tools to study and improve inulin content in chicory (Cichorium intybus). PhD thesis: Ghent University (Belgium).
Maroufi A. et al., 2012. Regeneration ability and genetic transformation of root type chicory (Cichorium intybus var. sativum). Afr. J. Biotechnol., 11, 11874-11886.
McKnight T.D., Lillis M.T. & Simpson R.B., 1987. Segregation of genes transferred to one plant cell from two different Agrobacterium strains. Plant Mol. Biol., 8, 439-445.
Miki B. & McHugh S., 2004. Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J. Biotechnol., 107, 193-232.
Murashige T. & Skoog F., 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plantarum, 15, 473-497.
Nandagopal S. & Kumari B.D.R., 2007. Phytochemical and antibacterial studies of chicory (Cichorium intybus L.). A multipurpose medicinal plant. Adv. Biol. Res., 1, 17-21.
Permingeat H.R. et al., 2003. Stable wheat transformation obtained without selectable markers. Plant Mol. Biol., 52, 415-419.
Puchta H., 2000. Removing selectable marker genes: taking the shortcut. Trends Plant. Sci., 5, 273-274.
Puchta H., 2003. Marker-free transgenic plants. Plant Cell Tissue Organ Cult., 74, 123-134.
Rakoczy-Trojanowska M., 2002. Alternative methods of plant transformation - a short review. Cell Mol. Biol. Lett., 7, 849-858.
Tuteja N. et al., 2012. Recent advances in development of marker-free transgenic plants: regulation and biosafety concern. J. Biosci., 37, 167-197.
Vervliet G. et al., 1975. Characterization of different plaque-forming and defective temperate phages in Agrobacterium. J. Gen. Virol., 26, 33-48.
Xin C. & Guo J., 2012. An efficient marker-free vector for clean gene transfer into plants. Afr. J. Biotechnol., 11, 3751-3757.
Xingguo Y. & Hua Q., 2008. Obtaining marker-free transgenic soybean plants with optimal frequency by constructing a three T-DNA binary vector. Front. Agric. China, 2, 156-161.
Zaidi M.A. et al., 2006. Optimizing tissue culture media for efficient transformation of different indica genotypes. Agron. Res., 4, 563-575.
Zubko E., Scutt C. & Meyer P., 2000. Intrachromosomal recombination between attP regions as a tool to remove selectable marker genes from tobacco transgenes. Nat. Biotechnol., 18, 442-445.
Zuo J., Niu Q.W., Moller S.G. & Chua N.H., 2001. Chemical-regulated, site-specific DNA excision in transgenic plants. Nat. Biotechnol., 19, 157-161.





