- Home
- Volume 14 (2010)
- numéro 4
- Gene expression and genetic analysis during higher plants embryogenesis
View(s): 6873 (25 ULiège)
Download(s): 163 (1 ULiège)
Gene expression and genetic analysis during higher plants embryogenesis

Editor's Notes
Received on November 21, 2009; accepted on March 30, 2010
Résumé
Analyse génétique et expression de gènes au cours de l'embryogenèse chez les plantes supérieures. Cette revue décrit et discute les récentes études ciblant l'analyse des processus de l'embryogenèse chez les plantes supérieures grâce à une combinaison d'approches descriptive, expérimentale et génétique. L'étude d'expression de gènes a permis de dégager des hypothèses sur les mécanismes induits au cours des phases précoces de l'embryogenèse chez les plantes supérieures. Ces mécanismes impliquent notamment une régulation transcriptionelle et post-transcriptionelle fine et spécifique et diverses voies de transduction du signal à chacun des stades de développement.
Abstract
This review describes and discusses recent attempts to analyze the embryogenesis process in higher plants, through combination of descriptive, experimental, and genetic approach. Analysis of gene expression profiles has permitted to build hypothesis concerning the induced mechanisms in early phases of embryogenesis in higher plants. Such mechanisms involve specific transcriptional and post-transcriptional regulatory pathways as well as diverse signal transduction processes at each stage of plant development.
Table of content
1. Introduction
1In flowering plant, embryogenesis begins with a double fertilization during which the two sperm cells of the pollen grain merge with the egg cell and the central cell to produce the zygote and the endosperm, respectively. The zygote then divides asymmetrically giving rise to two cells of different size and fate: the small apical cell forms the major part of the embryo proper whereas the big basal cell develops into the suspensor. Subsequently, the embryo grows according to two different axes, the apical-basal and the radial axes, to establish different organs and tissues (Umehara et al., 2007). This developmental switch involves not only differential gene expression, but various signal transduction pathways for activating/repressing numerous gene sets, many of which are yet to be identified and characterized. Several processes of flowering plant embryogenesis are now well known although limited genetic information is available during the early stages of seed development. Molecular and physiological events leading to the seed formation are still far to be completely understood. Mutagenic silencing of genes and characterization of model plant mutants such as Arabidopsis thaliana L. with abnormal embryo development have been used as genetic tool to understand gene regulation of plant embryogenesis (McElver et al., 2001). Several Arabidopsis mutants which are altered during early or late embryo development have been described and several genes have been identified that play a significant role during cell division and cell wall formation at various stages of embryo differentiation (Tzafrir et al., 2004). Moreover, large-scale single-pass sequencing of cDNA prepared from tissues with specific metabolism activity has provided an efficient and rapid mean towards isolation of new genes (Suh et al., 2003). The present review is thus an effort to collate information on various embryo-specific genes that have been identified during flowering plant embryogenesis. It also focuses on the potential benefits and limitations of mutants analysis in relation with plant embryo development. In particular, we discuss some of the processes involved in embryo formation and its genetic control; the signaling systems that regulate the expression of relevant genes are also examined.
2. Genetic analysis of plant embryogenesis
2The growth and development of higher plants are characterized by the execution of cell division, expansion and differentiation along two axes: the apical-basal and the radial axis. Genetic approach was developed to investigate embryogenesis development in plant by isolating and characterizing mutants exhibiting abnormal embryo development. These mutants represent an important tool to characterize the corresponding genes and to elucidate their roles. Analyses of seedling lethal mutants led to the hypothesis that early dicotyledonous embryo is divided into three domains: apical, central and basal (Laux et al., 2004). The apical embryo domain will generate the shoot meristem and most of the cotyledons; the central embryo domain will form the hypocotyl and roots and contribute to cotyledons and the root meristem; the basal embryo domain will give rise to the distal parts of the root meristem, and the stem cells of the central root cap. Two meristematic cell populations arise during embryogenesis: the Shoot Apical Meristem (SAM) generating the aerial part of the plant, and the Root Apical Meristem (RAM) generating the underground part.
2.1. The apical domain of the embryo
3Phytohormones such as auxin and several groups of genes cooperate to balance meristem maintenance and organ production (Figure 1). Mutations in genes involved in the apical embryo domain specification are predicted to disrupt both cotyledon and shoot meristem development. SHOOT MERISTEMLESS (STM) gene is required for SAM formation during embryogenesis and is expressed by only a specific set of cells within the embryo apex. STM has been shown to code for a class I KOTTED-like homeodomain containing protein. In the mutants stm organs originating from the shoot meristems are fused, which indicates a role of STM in restricting cells and organ formation (Long et al., 1998). The WUSCHEL (WUS) gene has been shown to encode homeobox genes family, which involves important regulatory genes in the speciation of cell fate and body plant at the early stage of embryogenesis in higher organism. Moreover, WUS and STM play complementary roles in maintaining shoot meristem. However, mutant wus and stm studies suggested that combined expression of both WUS and STM can trigger the initiation of ectopic meristem and organogenesis even in differentiated tissues (Lenhard et al., 2002). The cell differentiation is controlled by the genes CLAVATA (CLV). The CLV genes are required to limit the meristem size (DeYoung et al., 2008). CLV and STM genes are known to play opposite or competitive roles in the regulation of meristem activity. The interaction between the WUS and CLV pathways establishes a negative feedback loop between the stem cell and the meristematic regions. Indeed, WUS expression is sufficient to induce meristem cell identity, and expression of CLV genes represses meristem maintenance and WUS activity. The apical region of the globular embryo is partitioned into two domains: a peripheral domain of STM-expressing cells and a central domain showing expression of CUP-SHAPED COTYLEDON (CUC) genes. CUC genes regulate both the formation of the embryonic SAM and the separation of cotyledons and floral organs (Takada et al., 2001). Fujita et al. (2008) showed that embryo treatments with various polar auxin inhibitors stopped cotyledon and SAM development. This indicates the role of auxin signaling in the patterning of the apical embryo domain. Mutations in other homeobox genes families important for embryonic shoot meristem development, such as homeodomain LEUCINE ZIPPER (HD-ZIP) and homeodomain FINGER (PHD-FINGER) families reveal that these genes not only are required for stem cell initiation but also to maintain them. Functional analyses of zll mutants and gene expression indicate that ZWILLE (ZLL) is required for stem cell fate within the developing embryonic shoot meristem (Moussain et al., 1998). STM gene interacts with ZLL, which is expressed at the late globular stage in the apical domain. Moreover, ZLL gene appears to play a similar role as STM, and may be regulating the spatial expression pattern of STM (Moussain et al., 1998). GURKE (GK) is assumed to ensure correct organization of the shoot apical region (Torres-Ruiz et al., 1996). Indeed, mutants gk showed apical defects in seedlings but also reduced the efficiency of seed germination and seed growth. Likewise, the PASTICCINO (PAS) genes which are involved in the control of cell division and differentiation are required for normal organization of the apical region in the embryo (Faure et al., 1998). PASTICCINO mutants such as pas1 and pas2 produce abnormal embryos with altered cotyledon primordial leading to a flat apex (Bellec et al., 2002). More recently, Baud et al. (2004) identified lethal acc1 mutants affected in acethyl-CoA carboxylase 1 (ACCase 1) that displays a similar embryo phenotype to GURKE and PASTICCINO mutants. Genetic analyses have identified multiple gene functions involved in embryonic pattern; among them a member of the WOX genes family is differentially expressed in the small apical cell after the asymmetric division of the zygote. WOXs genes encode a WUSCHEL-related homeobox gene family. WOX2 has a putative zinc finger domain downstream of the homeodomain. Transcripts are expressed in the egg cell, the zygote and the apical cell lineage. This gene is necessary for cell divisions that form the apical embryo domain (Haecker et al., 2003). Previous studies have shown that STIMPY (STIP)/WOX9 are required for maintaining cell division and preventing premature differentiation in emerging seedlings. Complete loss of STIP activity results in early embryonic arrest, most likely due to a failure in cell division. STIMPY-LIKE (STPL)/WOX8 share redundant functions with a more related member of the WOX genes family such as WOX2, in regulating embryonic apical patterning (Kieffer et al., 2006). These findings show that combinatorial action of WOX transcription factors is essential for embryonic development. Mutation in LATERAL ORGAN BOUNDARIES (LOB) leads to alterations in size and shape of leaves and floral organs and causes male and female sterility (Shuai et al., 2002). Characterization of mutants lob suggests a potential role for this gene in organ separation or lateral organ development. Other genes required for shoot apical meristem formation are defined by ALTERED MERISTEM PROGRAM 1 (AMP1) which plays a role in balancing and restricting the meristem-promoting activity of auxin signaling. Mutants amp1 showed diverse morphological abnormalities including supernumerary cotyledons (Vidaurre et al., 2007). Characterization of amp1 and mp mutants suggested that AMP1 and MONOPTEROS (MP) antagonistically regulate embryo and meristem development in Arabidopsis.
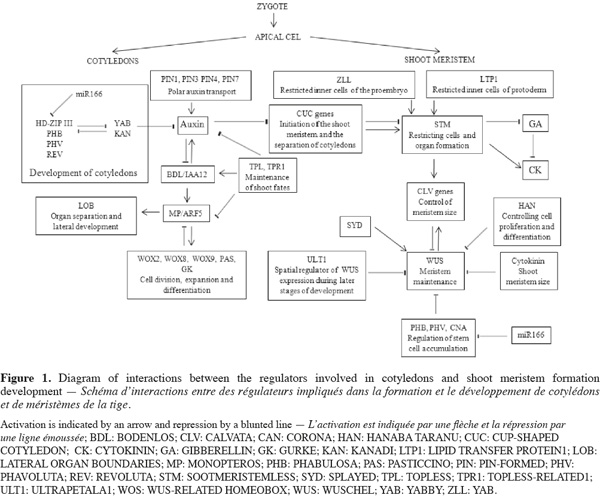
4To sustain the post-embryonic development of the shoot, it is crucial that the balance between the indeterminacy of the SAM and the determinate growth of lateral organs is maintained. The transcription factor of the homeobox-LEUCINE ZIPPER (HD-ZIP) proteins such as PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) play critical roles in this process (Prigge et al., 2005). It is likely that PHB, PHV, and REV regulate embryonic SAM formation by positively regulating STM expression. Mutants phb, phv, and rev display strongly reduced auxin polar transport to inflorescence, stems and hypocotyls. This alteration results in disrupted cell differentiation and morphology. A fundamental aspect of the transition phase of embryogenesis, when apical and basal organ fates are established, is the repression of root-promoting gene cascades in the apical embryo domain, which is provided by the activity of the TOPLESS (TPL) and TOPLESS-RELATED (TPR) proteins, co-repressors which function together with AUX/IAA proteins in auxin-mediated transcriptional repression (Szemenyei et al., 2008).
2.2. The central and basal domains of the embryo
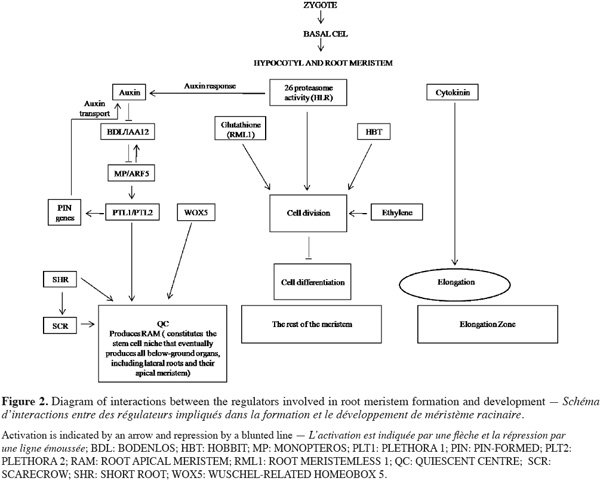
5Together the central and basal domains of the embryo contribute to RAM formation. Defects in specification signal in both regions disrupt organization of provascular cells. Both auxin and ethylene hormone express in the RAM. Auxin plays a conspicuous role in the specification and maintenance of stem cell and meristematic activity, while ethylene has been demonstrated to participate in the regulation of the cell division activity of the Quiescent Center (QC), a group of non dividing cells, which maintains the undifferentiated state of the surrounding stem cells. Alteration of the ethylene pathway by genetic or chemical tools affects the division activity of the QC suggesting its functions in maintenance of stem cell niche by regulating the balance between proliferation and quiescence of stem cells (Ortega-Martinez et al., 2007). It is now clear that activity of some regulator factors and normal auxin signalling are required for QC identity and thus for meristem maintenance and root growth (Figure 2). Studies of genes involved in auxin response and embryo proper development such as BODENLOS (BDL), MONOPTEROS (MP), and GNOM (GN), suggest that early root development depends on these gene activities. GN proteins are involved in the specification of apical-basal pattern formation and were recently considered as essential for cell division, expansion, and adhesion (Robert et al., 2008). Mutant gn embryos showed a loss of apical-basal polarity and alteration in radial pattern that displays abnormal intracellular traffic of transcription regulators (Mayer et al., 1993). Mutants bdl and mp are disrupted at the two-cell stage, when the apical cell divides horizontally rather vertically. These data suggest that MP and BDL are implicated in auxin-mediated apical-basal patterning process. The cells of the central domain divide abnormally and the hypophysis fails to produce the lens-shaped cell, resulting in seedlings devoid of the root, the root meristem, and the hypocotyls. MP and BDL are expressed only in embryo proper and are co-expressed in the early globular embryo; they can interact to form heterodimers (Hardtke et al., 2004). In contrast to MP and BDL, the first defects in AUXIN RESISTANT6 (AXR6) embryo are aberrantly oriented cell divisions in the derivatives of the basal daughter cell of the zygote; subsequently, no root meristem is formed and axr6 mutant seedlings are arrested in their development soon after germination (Hobbie et al., 2000). Mutants mp, bdl, and axr6 interfere to varying degrees with the formation of the embryo axis and are recognized by less oriented divisions in the basal domain. Contrarily to mutant mp gene, mutations in HOBBIT (HBT) gene affect the development of the basal, but not the central region, resulting in the lack of a functional root meristem, and therefore root formation. In contrast to mp seedlings, hbt seedlings initiate but do not develop adventitious roots from their hypocotyls because they are unable to form new root meristem. It is clear that root development and organogenesis require not only transcriptional hormonal signaling network but also key components regulators such as sterols which are the major components modulating the fluidity and permeability of eukaryotic cell membranes and in addition serve as precursor for steroid-signaling molecules that are known to elicit a variety of physiological processes (Kepinski, 2006). Functional roles for the campesterol-derived brassinosteroids (BRs) are established for integrating light signals in the regulation of postembryonic plant growth. Analyses of FACKEL (FK) activities indicate important roles of sterols for proper embryo development. Mutants fk provoke failure of cells in the central embryo domain fated to form vascular primordium. Later during development, mutant embryos also show abnormalities in the apical and basal domains and often form multiple shoot meristems and cotyledons (Schrick et al., 2000). Phenotypic abnormality is characterized by seedlings in which the hypocotyls is missing and by cotyledons attached directly to a short root. These data suggest that FK gene has the spatially restricted task of hypocotyls specification contrarily to MP and BDL genes characterized by mutants identified with impaired capacities for the production of both hypocotyls and root.
3. Genetic analysis of seed maturation
6Seed maturation is an important phase of seed development during which embryo growth ceases and storage products accumulate. The spatial and temporal regulation of all these processes requires the concerted action of several signaling pathways that integrate information from genetic programs, and both hormonal and metabolic signals (Figure 3).
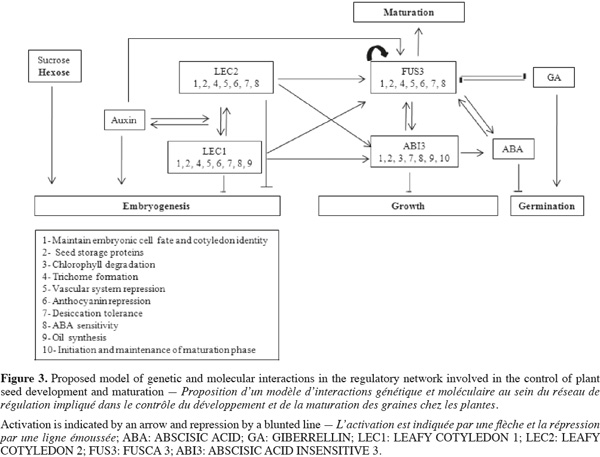
7The LEAFY COTYLEDON 2 (LEC2), LEAFY COTYLEDON 1 (LEC1), FUSCA3 (FUS3), and ABSCISIC ACID INSENSITIVE3 (ABI3) have been identified as master regulators of seed maturation (Santos Mendoza et al., 2005). Interestingly, study of abi3, fus3, lec1, and lec2 single, double and triple mutants show that these different regulators interact to control the various seed maturation features. Ectopic expression of LEC1 has demonstrated that this gene is required for the maintenance of the suspensor cell fate, the specification of cotyledon identity, the initiation and maintenance of the maturation phase, the promotion of embryonic cell identity and division, and the suppression of germination (Casson et al., 2006). Moreover, seed storage protein (SSR) gene expression is controlled by LEC1 through the regulation of ABI3 and FUS3 expression (Kagaya et al., 2005). FUS3 expression is regulated by LEC2 and FUS3 itself in the root tip, by LEC2 and ABI3 in the embryo axis, and by the four regulators (LEC1, LEC2, ABI3, and FUS3) in cotyledons. The expression of ABI3 is also controlled by the four regulatory proteins in cotyledons (Santos Mendoza et al., 2005). Indeed, both post-transcriptional and post-translational regulatory mechanisms are found to control ABI3 expression. However, ABI3 appears to be involved in auxin signaling as well as lateral root development. Additionally, ectopic expression of LEC2 suggests that auxin can induce this gene expression. Moreover, FUS3 expression is up-regulated by auxin in the embryo where it further coordinates ABA-GA action by feedback loops associated with Abscisic Acid (ABA) and Gibberillins (GA) synthesis (Gazzarini et al., 2004). Furthermore, FUS3 has been shown to regulate SSR gene expression by two ABA-independent mechanisms that seem different from that of ABI3 (Kagaya et al., 2005). Other important regulators for seed development and/or maturation have been identified (Gallardo et al., 2008). PICKEL (PKL) a chromatin-remodeling factor (CHD3) acts in concert with GA to repress embryonic traits during and after germination, including the expression of LEC1, LEC2, and FUS3 in roots (Li et al., 2005). Several pkl mutants which exhibit reduced GA transport cause alteration in embryo patterning. Yamagishi et al. (2005) isolated TANMEI (TAN) which controls various aspects of both early and late phases of embryo development. Analysis of tan mutants indicates that this gene plays a role in both the morphogenesis and maturation phases of embryogenesis. Moreover, tan mutants shares many characteristics with lec mutants, suggesting that TAN has overlapping functions with the LEC genes. For example, both tan-1 and lec mutations cause desiccation intolerance, defects in storage protein and lipid accumulation, and trichome formation on cotyledons. Together, these results indicate that TAN and LEC genes, particularly LEC1 and LEC2, interact synergistically. Moreover, TAN and LEC genes may operate in closely related pathway during embryogenesis (Yamagishi et al., 2005).
4. Embryo-defective mutants
8A vast number of genes represented by embryo-defective mutants may be required for normal embryogenesis, and they may be master regulators of this process (Table 1). Several embryo-lethal mutants in Arabidopsis have been identified and analyzed (McElver et al., 2001). These authors estimated that 750 genes can be easily mutated to give an embryo lethal phenotype. On the other side, Tzafrir et al. (2004) showed that 250 genes are essential to control normal phenotype in Arabidopsis seed development.
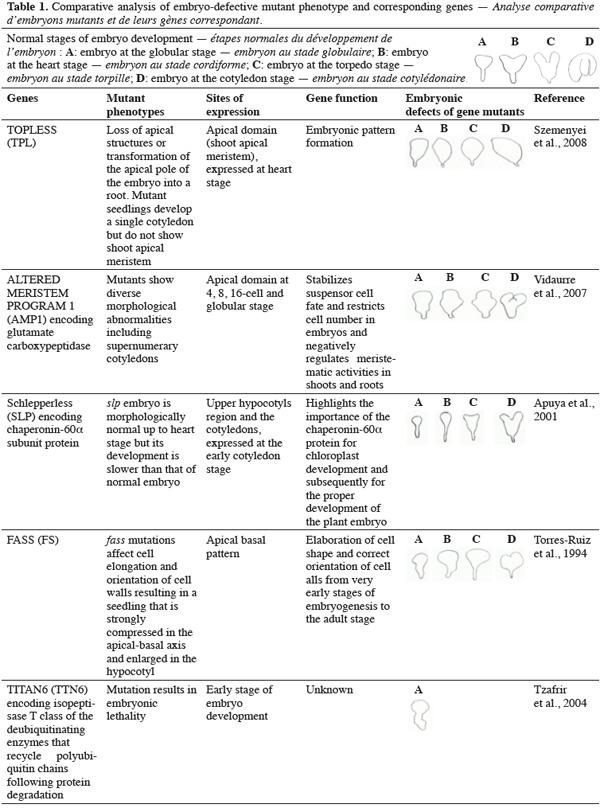
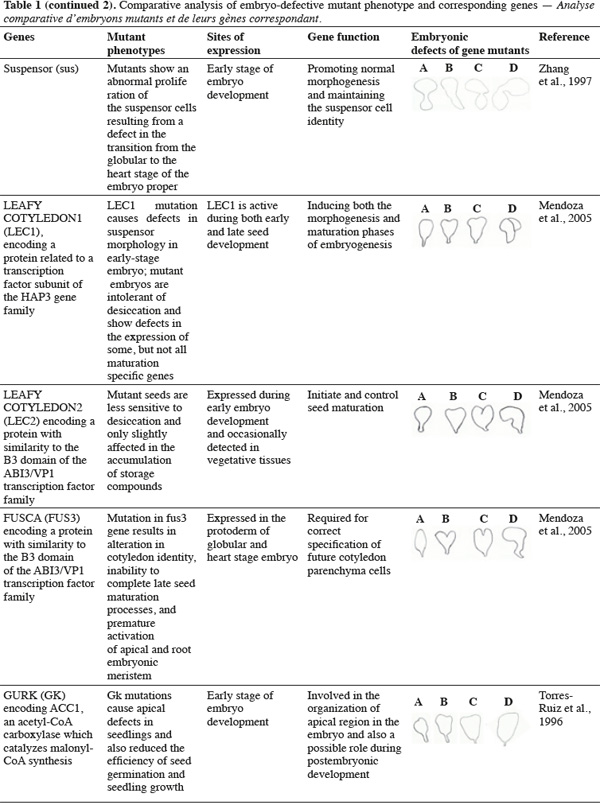
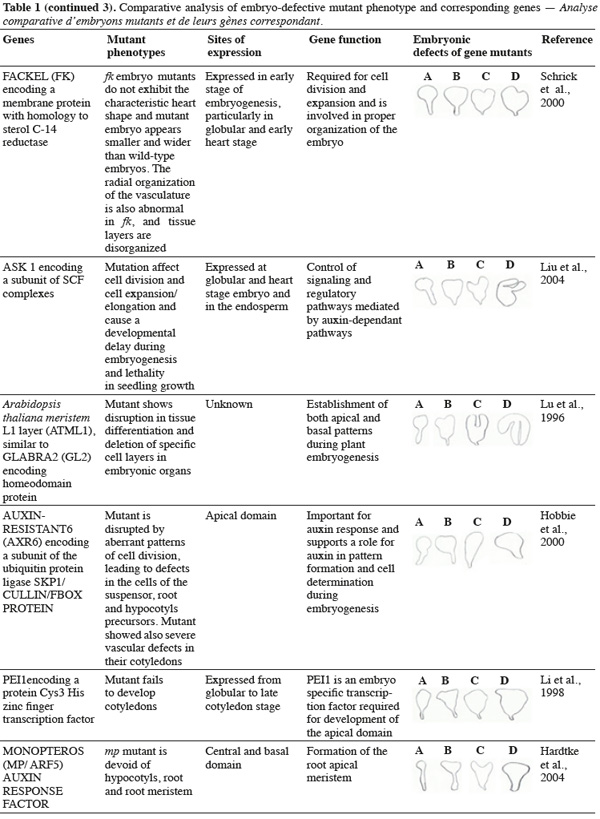
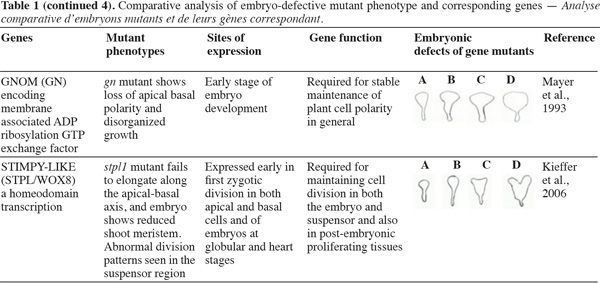
9Mutant study showed that disruption of the normally regular patterns of cell division in the Arabidopsis embryo does not necessarily interfere with proper cell fate determination. These data suggest that precise cell division is not required for pattern formation. Moreover, several embryo mutants show genetic disruptions for polar auxin transport and auxin response.
10Mutations in FASS (FS) gene produce disordered patterns of cell division. The fass mutants show inability to form microtubule preprophase bands, which predict the site of future cell wall deposition in plant cells (Torres-Ruiz et al., 1994). However the full range of appropriate cell types is present and complete in the fass mutants, albeit disordered and seedlings are produced. These data suggest that lineage-dependent segregation of cell fate determinants does not play a crucial role in patterning. Other mutants, such as Knolle (Kn), and Keule (Keu) also disrupt cell division and shape. At the cellular level, mutant embryos are characterized by incomplete cross walls and enlarged cells with polyploidy nuclei. KNOLLE (KN) encodes a member of the syntaxin family, a class of membrane-bound receptor required for docking and fusion of vesicles at the target membrane (Lukowitz et al., 1996), whereas KEULE (KEU) gene encodes a member of the Sec1 protein family which regulates vesicle trafficking and interacts with syntaxins (Assaad et al., 2001). Additional mutation in HINKEL gene, a Kinesin-like protein apparently involved in reorganization of phragmoplast microtubules (Strompen et al., 2002), show similar embryo-lethal phenotypes as KN and KEU genes.
11The Arabidopsis KIESEL (KIS) encodes Tubilin-Folding Cofactors (TFCs) involved in the control of the α/β-tubilin monomer balance which is important during microtubule biogenesis. In plant, microtubules have an important function in chromosome separation, cell division, and the establishment of cell polarity. Mutations in kis affect the biogenesis of microtubules and result in lethal embryos with one or few enlarged cells and few microtubules (Kirik et al., 2002).
12Several genes regulate a wide range of cellular processes in eukaryotes and their mutations result in severe cell division defects during endosperm and embryo development. The TITAN (TTN) genes encode chromosome scaffold proteins of the condensing and cohesion classes named STRUCTURAL MAINTENANCE OF CHROMOSOME (SMC) proteins (condensins and cohesions) required for chromosome function at mitosis. Development of ttn1 and ttn2 embryos is arrested at 1-2 cell stage, whereas ttn3 can produce fertile homozygous mutant plants. Embryo development is stopped shortly after fertilization in most ttn mutants and in some cases is accompanied by dramatic cell enlargement. Moreover, embryo growth is arrested in several mutants with defects in embryo and endosperm at different stages of embryo development (Liu et al., 1998).
13Disruption in RPN1 gene encodes subunit of the 26S proteasome (considered as negative regulators of auxin signaling for ubiquitin-mediated proteolyse) in Arabidopsis and provokes embryo lethality (Brukhin et al., 2005). The regulatory particle (RP) consists of 17 subunits, which form two subparticles, the lid and the base. The base contains three non-ATPase subunits (RPN1, RPN2, and RPN10) and six ATPase subunits (RPT1 to RPT6), which contact the α-subunit ring and likely assist in substrate unfolding and transport (Brukhin et al., 2005). The lid binds to the base and contains the other nine subunits (RPN3, RPN5 to RPN9, and RPN11 to RPN13). Embryos of rpn1 mutants stop their growth at the globular stage with defects in the formation of the embryonic root, the protoderm, and procambium. Mutant phenotypes analyses show that RPN12a subunit of Arabidopsis thaliana participates in cytokinin responses (Smalle et al., 2002). These data suggest the indirect mechanism of auxin action contrasting with the direct effects of cytokinin on bud growth; there is good evidence that auxin can regulate cytokinin biosynthesis.
14The ARP7 gene encodes actin-related proteins involved in chromatin dynamics and transcriptional regulation. A substantial knockdown of its expression severely affects plant architecture and considerably delays the abscission of floral organs. Mutants arp7 produce morphologically aberrant embryos that are arrested at or before the torpedo stage of development (Kandasamy et al., 2005). Genes that affect the function and/or development of organelles may also be playing similarly significant roles in embryogenesis. A mutation in EDD1 encodes a plastedic form of glycyl-tRNA synthase (Uwer et al., 1998), and leads to embryo arrest between globular and heart stages. On the other side, embryo mutants Schlepperless (slp), a plastid chaperonin-60α subunit protein involved in chloroplast development, show lesion in the chloroplast and delay of embryo development before heart stage. The embryonic cotyledons of this mutant are highly reduced (Apuya et al., 2001).
15Arrest of embryo growth accompanied by abnormal mitotic division is commonly observed in aborted seeds of some mutants such as TROMOZ (TOZ). Indeed, toz mutation affects cell division during embryogenesis and mutant embryos stop at globular stage (Griffith et al., 2007). Mutations severely disrupt cytokinesis (specific phases lead to cytoplasm partitions of a dividing cell and synthesis of a new plasma membrane) during embryo and endosperm formation (Sorensen et al., 2002). These data suggest that TOZ affects early embryonic development.
16In Arabidopsis thaliana, genes that regulate suspensor development have been isolated by studies of mutants, and the functions of these genes have been characterized (Zhang et al., 1997). Indeed in some mutants, suspensor ceases to grow at different stages of embryo development; this arrest is accompanied by abnormal activity of the suspensor.
17TWN2 and TWN1 are considered as genes involved not only for maintaining suspensor identity but also for regulating organogenesis in the embryo apex (Vernon et al., 2002). Mutant twn2 show degenerates at an early stage; however suspensor cells initiate embryonic development and eventually form one or more embryo. Mutant twn1 shows defects both in the suspensor and the cotyledons (Vernon et al., 2002). Similarly, the abnormal raspberry1 (rsy1) and raspberry2 (rsy2) are characterized by a morphologically abnormal embryo that fails to form axis and cotyledons, and have an enlarged suspensor (Yadegari et al., 1994). RSY3 gene is localized in chloroplasts, and it has been suggested that RSY3 protein is required for chloroplast differentiation and embryonic development (Apuya et al., 2002). The raspberry mutant rsy3 shows abnormality in the embryo proper and is arrested in the globular embryo stage of development. These data suggest that twn and rsy mutations are affected components of the apical-basal cell signaling pathway.
18Mutants with abnormal suspensors have been identified and explored in part through the analysis of embryo-defective mutants in Arabidopsis thaliana. Mutant suspensors (sus1, sus2, and sus3) are both larger and wider than normal suspensors and the embryo-proper consistently is arrested at the preglobular stage of development. According to Zhang et al. (1997), as rsy and sus mutations cause the arrest of the embryo-proper, it could be argued that the abnormal suspensor development in these mutants results in the absence of a viable embryo-proper.
5. Conclusion and perspectives
19Embryogenesis represents a key step of plant life cycle during which embryo develops the main structures of the future individual according to a well-defined morphological pattern.
20During embryo development, different post-transcriptional regulation mechanisms appear to be required to control the genetic programs induced at each stage.
21Although histological, morphological, molecular, and biochemical studies have provided descriptive information about embryogenesis, a mechanistic understanding of the processes that underlie embryonic development is lacking.
22Important clues about the process involved in embryo development may be obtained from analyses of embryonic mutant. The identification and characterization of embryo-defective mutants have been shown to be potentially useful in dissecting events in embryo development as a genetic approach.
23Differential gene expression is a commonly used approach for interpreting gene expression profiles during embryo development. Many genes involved in diverse processes of embryo development have already been characterized through a wide range of techniques such as differential screening approaches or mutational screening. Seed development proceeds through a series of spatially and temporally regulated gene expression steps. Elucidation of gene expression patterns associated with a specific stage of seed development is critical for understanding the molecular and biochemical events, which may be characteristic of that stage.
24Recently, very sensitive techniques adapted to the study of transcriptional activities of scarce biological tissues and allowing genome-wide gene expression analysis, such as in silico expressed sequence tags (EST) analysis, SAGE (Serial Analysis of Gene Expression) or microarrays technology, have been developed (Kasai et al., 2005). These techniques give the opportunity to obtain global gene expression patterns during seed development in flowering plant, particularly in Arabidopsis thaliana and a few other well-characterized plant species such as Helianthus annuus L., Medicago truncatula L., and Brassica napus L. (White et al., 2000).
25More recently novel factors affecting plant embryogenesis have been identified (Spartz et al., 2008); cross-linking between phytohormone and transcription factors is likely to display a part of embryogenesis. However, the mechanism of plant embryogenesis remains partially unclear. Particularly results of epigenetic approaches suggest that DNA methylation and chromatin modification are also important factors for plant embryogenesis (Xiao et al., 2006). Future studies will clarify and highlight the interaction of these factors, thereby revealing the entire embryogenesis regulatory mechanism. Therefore, holistic approaches such as the integration of genomics, proteomics, and metabolic, from the perspective of systems biology, have a great potential in revealing the interaction between different signaling cascades involved in triggering embryogenesis in flowering plant.
Bibliographie
Apuya N.R. et al., 2001. The Arabidopsis embryo mutant Schlepperless has a defect in the Chaperonin-60α gene. Plant Physiol., 126, 717-730.
Apuya N.R. et al., 2002. RASPBERRAY3 gene encodes a novel protein important for embryo development. Plant Physiol., 129, 691-705.
Assaad F.F., Huet Y., Mayer U. & Jurgen G., 2001. The cytokenesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J. Cell Biol., 152, 531-543.
Baud S. et al., 2004. Gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Reports, 5, 515-520.
Bellec Y. et al., 2002. Pasticcino2 is a protein tyrosine phosphatase-like involved in cell proliferation and differentiation in Arabidopsis. Plant J., 32, 713-722.
Brukhin V. et al., 2005. The RPN1 subnit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell, 17, 2723-2737.
Casson S.A. & Lindsey K., 2006. The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol., 142, 526-541.
DeYoung B.J. & Clarks E., 2008. BAM receptors regulate stem cell specification and organ development through complex interaction with CLAVATA signaling. Genetics, 180, 895-904.
Faure J.D. et al., 1998. The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development, 125, 909-918.
Fujita T. et al., 2008. Convergent evolution of shoot in land plants: lack of auxin polar transport in moss shoots. Evol. Dev., 10, 176-186.
Gallardo K., Thompson R. & Burstin J., 2008. Reserve accumulation in legume seeds. C.R. Biol., 331, 755-762.
Gazzarrini S. et al., 2004. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellins and abscisic acid. Dev. Cell, 7, 373-385.
Griffith M.E. et al., 2007. The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis. Plant Cell, 19, 2246-2263.
Haecker A. et al., 2003. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development, 131, 657-668.
Hardtke C.S. et al., 2004. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYLA. Development, 131, 1089-1100.
Hobbie L. et al., 2000. The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development, 127, 23-32.
Kagaya Y. et al., 2005. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol., 46, 399-406.
Kandasamy M.K., McKinney E.C., Deal R.B. & Meagher R.B., 2005. Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol., 138, 2019-2032.
Kasai Y. et al., 2005. 5'SAGE: 5'-end serial analysis of gene expression database. Nucleic Acids Res., 33, 550-552.
Kepinski S., 2006. Integrating hormone signaling and patterning mechanisms in plant development. Curr. Opin. Plant Biol., 9, 28-34.
Kieffer M. et al., 2006. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell, 18, 560-573.
Kirik V. et al., 2002. The Arabidopsis TUBULIN-FOLDING COFACTOR gene is involved in the control of the α/β-tubulin monomer balance. Plant Cell, 14, 2265-2276.
Laux T., Würschum T. & Breuninger H., 2004. Genetic regulation of embryonic formation. Plant Cell, 16, 190-202.
Lenhard M., Jürgen G. & Laux T., 2002. The WUSCHEL and SHOUTMERISTEMLESS gene fulfill complementary roles in Arabidopsis shoot meristem regulation. Development, 129, 3195-3206.
Li H.C. et al., 2005. PIKEL acts during germination to repress expression of embryonic traits. Plant J., 44, 1010-1022.
Li Z. & Thomas T.L., 1998. PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell, 10, 383-398.
Liu C.M. & Meinke D.W., 1998. The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J., 16, 21-31.
Liu F. et al., 2004. The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell, 16, 5-20.
Long J.A. & Barton M.K., 1998. The development of apical embryonic pattern in Arabidopsis. Development, 125, 3027-3035.
Lu P., Porat R., Nadeau J.A. & O'Neill S.D., 1996. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell, 8, 2155-2168.
Lukowitz W., Mayer U. & Jurgens G., 1996. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell, 84, 61-71.
Mayer U., Büttner G. & Jürgens G., 1993. Apical basal pattern formation in the Arabidopsis embryo: studies on the role of the gnom gene. Development, 117, 149-162.
McElver J. et al., 2001. Insertional mutagenesis of gene required for seed development in Arabidopsis thaliana. Genetic, 159, 1751-1763.
Mendoza M. et al., 2005. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Letter, 579, 4666-4670.
Moussain B. et al., 1998. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBRO J., 17, 1799-1809.
Ortega-Martenez O., Pernas M., Carol R.J. & Dolan L., 2007. Ethylen modulates stem cell division in the Arabidopsis thaliana root. Science, 317, 507-510.
Prigge M.J. et al., 2005. Class III homeodomain-leucine zipper gene family members have overlapping antagonistic, and distinct roles in Arabidopsis development. Plant Cell, 17, 61-76.
Robert S. et al., 2008. Endosidin1 defines a compartement involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl Acad. Sci. USA., 105, 8464-8469.
Schrick K. et al., 2000. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev., 14, 1471-1484.
Shuai B., Reynaga-Pena C.G. & Springer P.S., 2002. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol., 129, 747-761.
Smalle J. et al., 2002. Cytokinin growth responses in Arabidopsis involve 26S proteasome subunit RPN12. Plant Cell, 14, 17-32.
Sorensen M.B. et al., 2002. Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development, 129, 5567-5576.
Spartz A.K. & Gray W.M., 2008. Plant hormone receptors: new perception. Genes Dev., 22, 2139-2148.
Strompen G. et al., 2002. The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol., 12, 153-158.
Suh M.C. et al., 2003. Comparative analysis of expressed sequence tags Sesamum indicum and Arabidopsis thaliana developing seeds. Plant Mol. Biol., 52, 1107-1123.
Szemenyei H., Hannon M. & Long J.A., 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science, 319, 1384-1386.
Takada S., Hibara K., Ishiba T. & Tassaka M., 2001. The CUP-SHAPED COTYLEDON 1 gene of Arabidopsis regulates shoot apical meristem formation. Development, 128, 1127-1135.
Torrez Ruiz R.A. & Jürgens G., 1994. Mutation in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development, 120, 2967-2978.
Torres-Ruiz R.A., Lohner A. & Jurgens G., 1996. The GURKE gene is required for normal organization of the apical region of the Arabidopsis embryo. Plant J., 10, 1005-1016.
Tzafrir I. et al., 2004. Identification of gene required for embryo development in Arabidopsis. Plant Physiol., 135, 1206-1220.
Umehara M., Ikeda M. & Kamada H., 2007. Endogenous factors that regulate plant embryogenesis: recent advances. Jpn. J. Plant Sci., 1, 1-6.
Uwer U., Willmitzer L. & Altmann T., 1998. Inactivation of glycyl-tRNA synthase leads to an arrest in plant embryo development. Plant Cell, 10, 1277-1294.
Vernon D.M., Hannon M.J., Le M. & Forsthoefel N.R., 2002. An expanded role for the TWN1 gene in embryogenesis: defects in cotyledon pattern and morphology in the twn1 mutant of Arabidopsis (Brassicaceae). Annu. J. Bot., 88, 570-582.
Vidaurre D.P., Ploense S., Krogan N.T. & Berleth T., 2007. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development, 134, 2561-2567.
White J.A. et al., 2000. A new set of Arabidopsis expressed sequence tags from developing seeds. The metabolic pathway from carbohydrates to seeds oil. Plant Physiol., 124, 1582-1594.
Xiao W. et al., 2006. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell, 18, 805-814.
Yadegari R. et al., 1994. Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryo. Plant Cell, 6, 1713-1729.
Yamagishi K. et al., 2005. TANMEI/EMB2757 encodes a WD repeat protein required for embryo development in Arabidopsis. Plant Physiol., 139, 163-173.
Zhang J.Z. & Somerville C.R., 1997. Suspensor-derived polyembryony caused by altered expression of valyl-tRNA synthetase in the twn2 mutant of Arabidopsis. Dev. Biol., 94, 7349-7355.
To cite this article
About: Ghassen Abid
Univ. Liege - Gembloux Agro-Bio Tech. Unit of Tropical Crop Husbandry and Horticulture. Passage des Déportés, 2. B-5030 Gembloux (Belgium). E-mail: gabid@student.ulg.ac.be – Walloon Agricultural Research Centre. Department of Biotechnology. Chaussée de Charleroi, 234. B-5030 Gembloux (Belgium).
About: Jean-Marie Jacquemin
Walloon Agricultural Research Centre. Department of Biotechnology. Chaussée de Charleroi, 234. B-5030 Gembloux (Belgium).
About: Khaled Sassi
Institut National Agronomique de Tunisie. Department of Agronomy and Plant Biotechnology. Laboratory of Agronomy. Avenue Charles Nicolle, 43. TU-1082 Tunis-Mahrajène (Tunisia).
About: Yordan Muhovski
Walloon Agricultural Research Centre. Department of Biotechnology. Chaussée de Charleroi, 234. B-5030 Gembloux (Belgium).
About: André Toussaint
Univ. Liege - Gembloux Agro-Bio Tech. Unit of Tropical Crop Husbandry and Horticulture. Passage des Déportés, 2. B-5030 Gembloux (Belgium).
About: Jean-Pierre Baudoin
Univ. Liege - Gembloux Agro-Bio Tech. Unit of Tropical Crop Husbandry and Horticulture. Passage des Déportés, 2. B-5030 Gembloux (Belgium).





