Quantification of MBM adulteration in compound fertilizers and composts by NIRS
China Agricultural University (East Campus). College of Engineering. Box 191. CN-Beijing 100083 (P. R. China). E-mail: hanlj@cau.edu.cn
China Agricultural University (East Campus). College of Engineering. Box 191. CN-Beijing 100083 (P. R. China).
China Agricultural University (East Campus). College of Engineering. Box 191. CN-Beijing 100083 (P. R. China).
China Agricultural University (East Campus). College of Engineering. Box 191. CN-Beijing 100083 (P. R. China).
Abstract
The objective of this study was to demonstrate the feasibility of using near infrared reflectance spectroscopy (NIRS) to determine MBM content in compound fertilizers and composts. One hundred fourty adulterated compound fertilizer samples were prepared in the laboratory by mixing 4 types of compound fertilizers with 3 types of MBM randomly at different levels of 0.1%-10.0% (w/w). One hundred twenty adulterated compost samples were obtained by mixing 41 compost samples with 28 MBM at different levels of 3.0%-24.0% (w/w). NIRS calibration models were developed using the partial least squares (PLS) regression method. Results showed that the coefficients of determination for calibration (R2) and validation (r2) were 0.996 and 0.622, 0.988 and 0.722 for adulterated compound fertilizers and composts respectively. The ratios of prediction to deviation (RPD) were 8.84 and 1.87 for them respectively. These results indicated that NIRS could be used to quantify the adulteration of banned MBM in compound fertilizers with high prediction accuracy, and be insufficient to determine the content of MBM in composts due to low prediction accuracy.
1. Introduction
1Feed contaminated with meat and bone meal (MBM) is commonly accepted as the main transmission carrier of the prion responsible for bovine spongiform encephalopathy (BSE). Since the first BSE case in the UK cattle herd in 1986 and its association with the variant Creutzfeldt-Jakob disease (CJD) in humans in 1996, processed animal proteins, including MBM, are banned for use as a feed ingredient for animals, and it is an important measure to prevent the spread of transmissible spongiform encephalopathies (TSE) (Gizzi et al., 2004). Meanwhile, use of compound fertilizers or composts adulterated with banned MBM in livestock grazing systems may cause potential BSE risk through the feed chain. The UK and Japan have prohibited the mammalian MBM from all animal feeds and fertilizers (Matibag et al., 2005).
2Although the processed animal protein including MBM was banned to use, some merchants are still driven by economic benefits for the production and sale of MBM. Therefore it is important for quantitative analysis of MBM in feeds, compound fertilizers, composts, or the other products. Several methods have been developed to detect ruminant tissue in feeds including classical microscopy, polymerase chain reaction (PCR), immunological methods (Ansfield et al., 2000; Engling et al., 2000; Bellagamba et al., 2001). However, these methods are costly and/or time-consuming, are often defeated by thermal damage to protein and DNA, and are inappropriate for routine testing of the feed tonnage trade globally (Murray et al., 2001).
3Near infrared reflectance spectroscopy (NIRS) is an attractive alternative, which is rapid and cost efficient, without the destruction of samples and the use of hazardous chemicals. Over the last few years, NIRS technology has been introduced in agriculture, food, petrochemical, textile and pharmaceutical fields (Williams et al., 2001), and some studies showed that it is feasible to quantify the MBM content in feeds by NIRS (Pérez-Marin et al., 2004). Amari et al. (2004) reported that the NIRS technique could also quantitatively analyze the MBM content in compound fertilizers. The objective of this study was to improve the study of using NIRS to determine MBM content in compound fertilizers and demonstrate the feasibility of MBM detection in composts.
2. Material and methods
2.1. Preparation of samples
4Four types of chemical compound fertilizers with different proportions of N, P and K, 15-15-15, 15-10-5, 14-11-8 and 12-8-5 respectively, were purchased in the rural markets in Beijing. Forty-one animal manure composts with different compositions, processing techniques and storage periods, were collected from farms and compost factories in Hebei Province and Beijing in China. Twenty-eight MBMs were obtained, 20 were from domestic mills and 8 were imported. All of them were crushed to pass a 1mm sieve. One hundred forty adulterated compound fertilizer samples were prepared in the laboratory by mixing the 4 types of compound fertilizers with 3 types of MBM randomly at different levels of 0.1%-10.0% (w/w). One hundred twenty adulterated compost samples were obtained by mixing 41 compost samples with 28 MBM at different levels of 3%-24% (w/w). All of the samples were stored at + 4 °C in a refrigerator until analysis by NIRS.
2.2. NIRS scanning
5A NIRS system SPECTRUM ONE NTS (Perkin Elmer, USA) was used for NIRS scanning. Samples were scanned in rotating quartz cells at ambient temperature. Each of the samples was scanned 3 times as log 1/R over the wavelength range 1,100 nm to 2,500 nm and the average spectrum was recorded prior to analysis.
2.3. Mathematical and statistical analysis
6Data analysis was performed using the Spectrum QUANT+ software provided by Perkin-Elmer. Preliminary data evaluation was performed using two parameters, spectral leverage and absolute residual of actual value, to test the outlier. Then samples were divided into a calibration set and a validation set based on concentration gradients. The fertilizer samples were sorted by the concentrations, and one of three removed as a validation sample, and the number of samples was 91 and 47 for calibration and validation set. For the compost samples, one of four removed as a validation sample, and the number of calibration and validation samples was 89 and 29, respectively. The NIRS calibration models were developed using the partial least squares (PLS) regression method. Various spectral regions and mathematic pre-treatment methods, including smoothing, derivative, standard normal variate (SNV) with detrending or without detrending and multiplicative scatter correction (MSC), were applied (Murray et al., 2001). Cross validations were also conducted to select the optimal number of factors and avoid overfitting (Shenk et al., 1995). The optimum spectral region and pre-treatment methods were selected according to the lowest standard error of cross validation (SECV), and the number of factors recommended by the software was adopted, which was determined by considering the lowest SECV with the fewest terms. The correlation coefficient of calibration (R2), the standard error of calibration (SEC), the correlation coefficient of determination (r2), the standard error of prediction (SEP) and the ratio of prediction to deviation (RPD, SD/SEP) were used to evaluate the calibrations. For RPD, the calibration is considered not to be useful below 1.5. Between 1.5 and 2.0, there is only a possibility of distinguishing between high and low values. From 2.0 to 2.5, approximate quantitative predictions can be made. Between 2.5 to 3.0, the quantitative prediction ability is good, because the prediction error is reduced to less than half of the error made when using the mean composition. Above 3.0, the prediction error is reduced by a factor of more than three and calibrations are considered of value for quality control (Williams et al., 1992; Saeys et al., 2005).
3. Results and discussion
3.1. MBM content in compound fertilizers and composts
7The mean, minimum and maximum values, and standard deviation (S.D.) of the MBM content in compound fertilizers and composts are summarized in table 1. The S.D. was 3.58 and 6.90 for adulterated fertilizer and compost samples respectively.
8Table 2 shows the statistics of calibration set and validation set for adulterated fertilizer and compost.
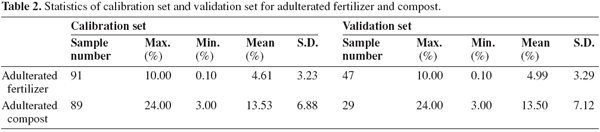
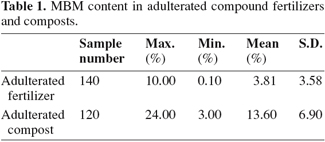
3.2. Sample spectra
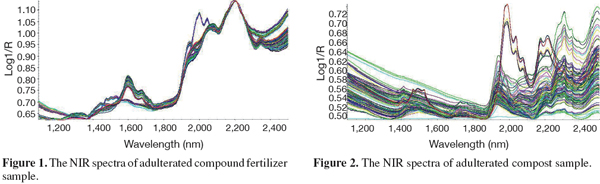
9Figures 1 and 2 show the NIR spectra of adulterated fertilizer and compost samples respectively. Representative spectra of the fertilizer, compost and MBM samples collected in the study are presented in figure 3. The rather strong absorption peaks of the representative spectrum of compound fertilizers appeared near the site of 2,004 nm, 2,047 nm and 2,196 nm et al. They were observed near the site of 1,935 nm, 2,060 nm, 2,211 nm, 2,278 nm and 2,308 nm et al. for compost samples, and near the site of 1,504 nm, 1,731 nm, 1,944 nm, 2,058 nm, 2,183 nm, 2,310 nm and 2,352 nm for MBM samples. These peaks are mainly caused by the overtone and combination absorptions of C-H, O-H and N-H stretching and deformation movements (Yan et al., 2005). Because there were substantial differences of organic chemicals in compound fertilizers, composts and MBMs, the position and intensity of absorption peaks were different accordingly in their spectra.
3.3. Outlier test and abnormal sample elimination
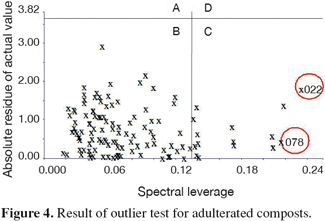
10Figure 4 shows the result of the outlier test for adulterated compost samples. The outlier samples in zone D could be deleted directly. The abnormal samples in zone A and zone C should not be readily removed but be analyzed or scanned again, and it would be deleted if still abnormal. For adulterated compost samples there were no samples located in zone A or D. The most outer samples were spectral abnormal in zone C, and only two samples (n°022 and 078) were finally removed, for their omission would bring in the effective decrease of the SEP value.
3.4. NIR calibrations and validations
11The optimum spectrum regions, pre-treatment methods and statistics of the calibrations and validations for compound fertilizers and animal manure composts are given in table 3.
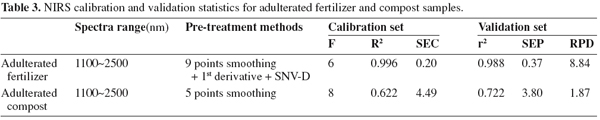
12The pretreatment method of 1st order derivative in conjunction with 9 points smoothing and SNV-D gave the lowest SEP values for adulterated fertilizer samples, and so did the 5 points smoothing method for adulterated compost samples. In this study, we found that the methods with 2nd order derivative generally performed poorly and brought higher SEP values. This may be due to the shortcoming of 2nd order derivative of easily sharpening small interfering information. Figure 5 displays the scatter plots of NIRS prediction versus actual values of the MBM contents in adulterated fertilizer and compost samples for the calibration and validation sets.
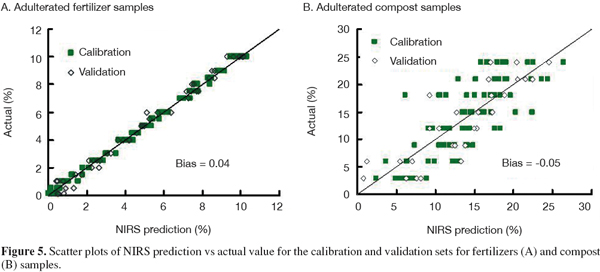
13For adulterated fertilizer samples with MBM, both of the R2 (0.996) and r2 (0.988) were greater than 0.95, the SEC and SEP values were rather low at 0.20% and 0.37% respectively. Its RPD value (8.84) was greater than 3.0, which suggests that the prediction error is reduced by a factor of more than three and calibrations were considered excellent. Comparisons of the R2 and r2, SEC and SEP show that the differences between calibration and validation were rather low, indicating the prediction was good. The RPD value of the calibration obtained in the present paper was 8.84, which compared well with that reported by Amari et al (2004), that is RPD = 15.10.
14Concerning the adulterated manure compost samples with MBM, the results were not as good as for the adulterated fertilizer samples. The R2 and r2 values were 0.622 and 0.722, and the SEC and SEP values were a little high at 4.49% and 3.80% respectively. The RPD value was lower than 2.5, which indicated the calibration established in this study did not perform well in order to accurately determine the content of MBM in composts and merely had the ability of distinguishing compost samples of high and low content of MBM. This might be due to the great diversity in the compost samples used to build the calibration. The samples collected in this study covered a wide area of composting materials, techniques and storage periods. The complicated spectral background suggests that further work is needed to build a robust library from which to develop more accurate calibrations. The use of non-linear algorithms as has been recommended by Pérez-Marín et al. (2005) for the prediction of ingredients in compound feeds could bring out a further improvement in the equation performance.
4. Conclusion
15Based on 140 adulterated compound fertilizer and 120 adulterated compost samples of local market, the present study demonstrated that NIRS could be used to quantify the adulteration of banned MBM in compound fertilizers with high prediction accuracy, and be insufficient to determine the content of MBM in composts due to a low prediction accuracy.
16Further work is needed to build robust models both for fertilizers and compost which can accommodate the huge diversity of samples encountered in the field and in the market.
17Acknowledgements
18The authors gratefully acknowledge the NIRS instrument assistance of Professor Shungeng MIN (China Agricultural University), and special thanks go to Masahiro AMARI (Japan National Institute of Livestock and Grassland Science) for his helpfulness and expert suggestions.
Bibliographie
Amari M., Otani F. & Matsumoto M., 2004. Quantitative analysis of MBM in fertilizer by NIRS. Grassl. Sci., 50(suppl.), 442-443.
Ansfield M., Reaney S.D. & Jackman R., 2000. Performance assessment and validation of a sensitive immunoassay for detection of ruminant and porcine heat stable proteins in compound animal feedstuffs. Food Agric. Immunol., 12(4), 285-297.
Bellagamba F., Moretti V.M., Comincini S. & Valfre F., 2001. Identification of species in animal feedstuffs by polymerase chain reaction-restriction fragment length polymorphism analysis of mitochondrial DNA. J. Agric. Food Chem., 49, 3775-3781.
Engling F.P., Jorgenson J.S., Paradies-Severin I. & Hahn H., 2000. Evidence of animal meal in feeds. Kraftfutter Feed Mag., 1, 14-17.
Gizzi G. et al., 2004. Determination of processed animal proteins, including meat and bone meal, in animal feed. J. AOAC Int., 87(6), 1334-1341.
Matibag G.C., Igarashi M. & Tamashiro H., 2005. BSE safety standards: an evaluation of public health policies of Japan, Europe, and USA. Environ. Health Prev. Med., 10, 303-314.
Murray I., Aucott L.S. & Pike I.H., 2001. Use of discriminant analysis on visible and near infrared reflectance spectra to detect adulteration of fishmeal with meat and bone meal. J. Near Infrared Spectrosc., 9, 297-311.
Pérez-Marin D.C., Garrido-Varo A., Guerrero-Ginel J.E. & Gómez-Cabrera A., 2004. Near-infrared reflectance spectroscopy (NIRS) for the mandatory labelling of compound feedingstuffs: chemical composition and open-declaration. Anim. Feed Sci. Technol., 116, 333-349.
Pérez-Marín D.C., Garrido-Varo A., Guerrero-Ginel J.E. & Gómez-Cabrera A., 2005. Implementation of local algorithm with NIRS for compliance assurance in compound feedingstuffs. Rev. Appl. Spectrosc., 59(1), 69-77.
Saeys W., Mouazen A.M. & Ramon H., 2005. Potential for onsite and online analysis of pig manure using visible and near infrared reflectance spectroscopy. Biosystems Eng., 91(4), 393-402.
Shenk J.S. & Westerhaus M.O., 1995. Analysis of agriculture and food products by near infrared reflectance spectroscopy. Silver Spring, MD, USA: NIRSystems.
Williams P.C. & Sobering D., 1992. In near infrared spectroscopy: bridging the gap between data analysis and NIR applications. In: Hildrum K., Isaksson T., Naes T. & Tandberg A., eds. Proceedings of the 5th ICNIRS, Haugesund, Norway. Chichester, UK: Ellis Horwood Ltd, 441-446.
Williams P. & Norris K., 2001. Near-infrared technology in the agricultural and food industries. 2nd ed. Saint Paul, MN, USA: American Association of Cereal Chemists.
Yan Y.L., Zhao L.L., Han D.H. & Yang Z.M., 2005. The chemical basis for near-infrared spectra analysis. In: Wang C. et al., eds. The technique and application for near-infrared spectra analysis. 1st ed. Beijing, China: China Light Industry Press, 29-39.

