Isolation and screening of Weissella strains for their potential use as starter during attiéké production
Received on October 4, 2014; accepted on April 13, 2016
Résumé
Isolement et sélection de souches de Weissella comme de potentiels ferments pour la production d’attiéké
Description du sujet. La fermentation non contrôlée de l’attieké, effectuée à partir d’inocula traditionnels, est responsable de la grande variabilité sensorielle observée pour cette denrée alimentaire.
Objectifs. La présente étude vise à sélectionner des micro-organismes aptes à être utilisés comme des ferments microbiens pour une fermentation contrôlée de la pâte de manioc en attieké.
Méthode. Trois bactéries lactiques isolées de ferments traditionnels sont identifiées comme étant Weissella cibaria ou Weissella confusa par la technique de désorption-ionisation laser assistée par matrice couplée au spectromètre de masse à temps de vol (MALDI-TOF MS) et leurs propriétés technologiques mises en évidence in vitro.
Résultats. Les trois isolats de Weissella induisent une baisse du pH initial (principal facteur de sécurité alimentaire) de 6,2 du bouillon MRS (de Man, Rogosa and Sharpe) à une valeur en dessous de 4,2 en moins de 42 h. En plus de présenter une activité alpha-amylase, ces bactéries fermentent le raffinose, un sucre indigeste. Toutes les trois sont osmotolérantes. Par ailleurs, les isolats Wc 114 et Wc 69 sont thermotolérants, tandis que l’isolat Wc 114 présente une activité pectinase nécessaire au ramollissement de la pâte de manioc.
Conclusions. Les trois bactéries lactiques isolées (Wc 69, Wc 21 and Wc 114) présentent des propriétés technologiques intéressantes susceptibles de convenir pour contrôler et standardiser la qualité de l’attieké.
Abstract
Description of the subject. Variability observed in sensory characteristics of “attiéké” results from an uncontrolled fermentation process due to the use of artisanal starters.
Objectives. This study aims to screen microbial strains for their use during fermentation of cassava dough into attieké.
Method. Technological properties of three lactic acid bacteria (LAB) isolated from artisanal starters were highlighted in vitro, and these LAB were identified as either Weissella cibaria or Weissella confusa by Matrix Assisted Laser Desorption Ionisation-Mass Spectrometry (MALDI-TOF MS).
Results. The three Weissella isolates Wc 69, Wc 21 and Wc 114 induced in less than 42 h a decrease below 4.2 (main food safety factor) of the initial pH of MRS (de Man, Rogosa and Sharpe) broth. These strains are osmotolerant, present alpha-amylase activity and ferment the indigestible sugar raffinose. Moreover isolates Wc 114 and Wc 69 are thermotolerant, while Wc 114 presents a pectinase activity necessary for cassava dough softening.
Conclusions. Considering their technological properties, the three isolated Weissella strains could be suitable in optimizing and standardizing the quality of attieké.
1. Introduction
1Cassava is the most important staple crop in Africa, with over 57 million tons of roots and their processed products are more consumed than corn with 30 million tons (Diallo et al., 2013). In Côte d'Ivoire, attiéké or cassava couscous is the major fermented plant food (Kouamé et al., 2012). In recent years, there has been a change from its familial production to commercial production stimulated by urbanization as well as external demand due to exports (Akely, 2012). The quality of cassava-derived foods is largely dependent on a combination of processing technologies such as peeling, boiling, steaming, pounding, slicing, grating, roasting, soaking, pressing and fermentation. During attiéké production, producers use an artisanal starter obtained by fermenting cassava roots for three days. These traditional inocula constitute the main source of microorganisms involved in later cassava dough fermentation (Koussemon et al., 2010). Differences in these traditional starters induce variable organoleptic qualities of “attieké”. Fermentation of cassava dough by bacteria and yeasts does not only enhance detoxification; it also improves food quality and safety by product preservation, flavor development, cyanide reduction and changes in functional properties (Obilie et al., 2003; Padonou et al., 2010a). Lactic acid bacteria (LAB) were shown as predominant microorganisms during natural fermentation of cassava roots (Kostinek et al., 2007). From a quality and safety perspective, the use of LAB cultures as starters induces a rapid acidification of the cassava dough which inhibits pathogenic bacteria growth and prevents spoilage (Holzapfel, 2002). The characterization of microbial starters for control the cassava dough fermentation in Côte d’Ivoire has not been reported. Therefore, this study reports the technological properties of three LAB strains isolated from traditional cassava starters, identified as Weissella cibaria or Weissella confusa by MALDI-TOF MS.
2. Materials and methods
2.1. Selection of best acid producer strains
2The biological material used was constituted by “Adjoukrou” traditional cassava inocula collected in small-scales production of attieké from thirteen locations in Abidjan. These inocula were naturally grown from boiled cassava roots packed in old fermenting bags and left to ferment for three days. The samples collected were wrapped in plastic bags (Stomacher, Laboratoire Humeau, Rennes, France) and transported on ice to the laboratory within 1 h, where they were mixed and then subdivided in five aliquots. Preparation of stock solutions and inoculation of agar plates were carried out according to Coulin et al. (2006). Total viable lactic acid bacteria (LAB) counted in each sample were analyzed by spread plating the tenfold diluted samples into MRS (de Man, Rogosa and Sharpe), BEA (Bile Esculin Azide), MSE (Mayeux, Sandine and Elliker), M17 agar plates (all from OXOID, Basingstoke, Hampshire, UK), to obtain the widest possible variety of LAB associated with fermenting traditional cassava starters. The media were supplemented with nystatin (1%) to inhibit fungal growth. Petri dishes were then incubated in an anaerobic jar at 30 °C for 48 h. LAB were identified as Gram positive, oxidase and catalase negative, respectively. The isolates were purified on MRS agar plates, examined by electron microscope (magnification 1000x) and those with a rod shape (bacilli) were stored at -80 °C in MRS buffer medium supplemented with 20% (v/v) glycerol. The isolates were anaerobically tested at 30 °C over three days for their capacity to produce acid in MRS agar plates (on Petri dishes) containing 1% CaCO3. Acid producing strains were identified by clear areas surrounding the colonies; strains presenting relatively wide areas were selected. To confirm these results, a second screening was done as previously described by Dicks et al. (1987) with slight modifications. Each strain was inoculated in 5 ml MRS agar medium without beef extract, and with 0.004 g·l-1 of bromocresol purple in tubes. Acid production was monitored by formation of a yellow area in the tube and acidification capacity was analyzed throughout the visual scale of the yellow area’s spread. In parallel, aerobic and anaerobic growths were assessed using two inoculated tubes containing 5 ml of MRS broth supplemented with 0.004 g·l-1 of bromocresol purple. One of them was incubated aerobically at 30 °C, and the other one anaerobically in an anaerobic jar. The color change of the MRS broth from purple to yellow was used as growth indicator. The heterofermentative or homofermentative character was assessed through CO2 gas production capacity in MRS agar broth. An isolate was considered a homofermentative lactic acid producer if no gas was produced.
2.2. Assay of acidification capacity of isolates
3Strains selected as good acid producers were tested during 42 h for their capacity to reduce the pH below 4.2. For this purpose, MRS broth adjusted to pH 6.5 before autoclaving (pH 6.2 after autoclaving) was inoculated with 0.2% of an overnight preculture (OD600 = 1) and cultures were grown aerobically at 37 °C. Acid production was determined by measuring the culture pH after 6, 12, 18, 24, 30, 36 and 42 h. MRS broth medium for all acid production tests was prepared from a single batch and then spread into tubes of 5 ml before autoclaving. During microbial growth, the pH of MRS broth media was automatically measured by a pH-meter. Biomass evolution was analyzed through absorbance at 600 nm. The diluted supernatant was used for the quantification of residual glucose, L-lactate and titratable acidity. Concentrations of glucose (g·l-1) and L-lactate (g·l-1) were measured by using a glucose and lactate automatic analyzer YSI2700S ELECT (Yellow springs Instruments Co., Inc.) equipped with two membranes, one for glucose (2365) and another for L-lactate (2329). Total titratable acidity was determined by titrating the sample with 0.1N NaOH using phenolphthalein as indicator and the result was expressed as percentage of lactic acid. Strains able to induce a change of MRS broth pH from 6.2 to less than 4.2 at the end of the fermentation were selected for further screening studies.
2.3. Screening of enzymatic activities of isolates
4For alpha-amylase or pectinase production, LAB isolates were grown on modified MRS agar plates containing 20 g·l-1 cassava starch or pectin as the sole carbon source. The plates have been incubated at 48 °C for 24 h and then flooded with iodine. Production of amylase or pectinase was evident by a clear area surrounding the colonies. The β-glucosidase activity was tested using 4-nitrophenyl- β-D-glucopyranoside as substrate. The medium was prepared by adding 0.1 g of 4-nitrophenyl- β-D-glucopyranoside (Merck, Darmstadt, Germany) to 100 ml of 0.666 M NaH2PO4 (pH 6) (Merck, Darmstadt, Germany). The mixture was dissolved and filter-sterilized (millipore filter of 0.2 microns). The culture was grown on MRS agar in tubes for 24 h at 30 °C. Colonies were picked from the plates using a sterile loop and were emulsified in physiologic saline to McFarland Turbidity Standard No. 3. Thereafter, 0.75 ml of culture was added to 0.25 ml of the test medium for overnight. The culture was then stopped by adding 1 ml of 1M sodium carbonate solution. Positive isolates that produced β-glucosidase degraded the linamarin analogue and changed the color of the mixture from colorless to a distinct yellow.
2.4. Identification of isolates
5The fermentation profile of the strains was determined during 48 h with the API 50 CHL test kit (Biomerieux, Marcy-l’Etoile, France). Microbial identification was achieved by using APILAB Plus software version 3.3.3 from BioMerieux. Furthermore, the identification was also done by MALDI-TOF MS (Matrix Assisted Laser Desorption Ionisation-Time Of Flight Mass Spectrometry) technique according to Doan et al. (2012), which comprises three steps: sample preparation, MALDI-TOF MS analysis and MALDI-TOF MS data analysis. For sample preparation, bacterial cells were grown on MRS under aerobic conditions for 48 h at 28 °C and subsequently subcultured twice. Cell extraction for MALDI-TOF MS analysis was performed according to the formic acid and acetonitrile extraction described by Freiwald et al. (2009). Supernatant containing the extracted proteins was used for MALDI-TOF MS analysis. One μL of freshly prepared cell extract was spotted on a 384 Opti-TOF stainless steel MALDI target plate (AB Sciex, Nieuwerkerk aan den Ijssel, The Netherlands). Cell extracts were dried at room temperature. One μl of a 0.5% (w⁄v) α-cyano-4-hydroxycinnamic acid (α-CHCA) in 50:48:2 acetonitrile, water, trifluoroacetic acid solution was added and allowed to dry. For the MALDI-TOF MS analysis, bacterial fingerprints were acquired using the 4800 Plus MALDI TOF⁄TOFTM-Analyzer (Applied Biosystems, Framingham, MA, USA) in linear positive ion mode. Ions were generated by a 200-Hz-tripled UV Nd:YAG laser, accelerated in a 20-kV electric field through a grid at 19.2 kV and separated according to their m⁄ z ratio in a 1.5-m long linear field-free drift region. For each spot, 40 laser shots at 50 random positions within the spot were collected, automatically generating mass spectra in the mass range from 2,000 to 20,000 Da. A maximum laser intensity of 5,200 a.u. resulted in signal intensities varying between 5.0 × 102 and 1.0 × 104. Prior to the analyses, calibration was performed with the protein calibration standard I that includes insulin (m/z 5734.6), ubiquitin I (m/z 8565.9), cytochrome c (m/z 12361.5) and myoglobin (m/z 16952.3) supplemented with the adrenocorticotropic hormone (ACTH) fragment 18–39 MALDI-MS standard (m/z 2465.7) (Bruker Daltonics GmbH, Leipzig, Germany). The MALDI-TOF MS data analysis was performed by raw data (mass spectra) extracted as t2d files from the 4800 plus MALDI TOF⁄TOFTM-analyzer. The t2d files were imported in the Data Explorer 4.0 software (Applied Biosystems) and transformed to text files. These text files were imported into the BioNumerics 6.0 software package (Applied- Maths, Sint-Martens-Latem, Belgium) and converted to fingerprints for further analysis. Pearson’s product moment correlation coefficient was used to determine similarity between the spectra. Spectra were subsequently clustered using Unweighted Pair Group Method with Arithmetic Mean. BioNumerics was used to set up a database composed of the MALDI-TOF MS profiles of reference strains and isolates. From these experiments, three heterofermentative strains acting as fast acid producers on glucose and identified by MALDI-TOF MS as Weissella cibaria/confusa (Pearson correlation > 99%) were selected.
2.5. Behavior of isolates to environmental conditions
6Growth of the isolates was determined at 15, 30, 37, 45 and 55 °C; 0.5 and 1% lactic acid; 2, 5, 7 and 9% (w/v) NaCl; pH of 3, 5, 7 and 9. The basal medium was adjusted with 1M phosphoric acid and 1M NaOH and the tubes were incubated at the specific temperatures (for effect of temperature) at 37 °C for effect of pH, concentrations of lactic acid and NaCl. At the end of fermentation, growth was evaluated by visual inspection. All experiments were done in triplicate.
3. Results
3.1. General characteristics and tentative identification of isolates
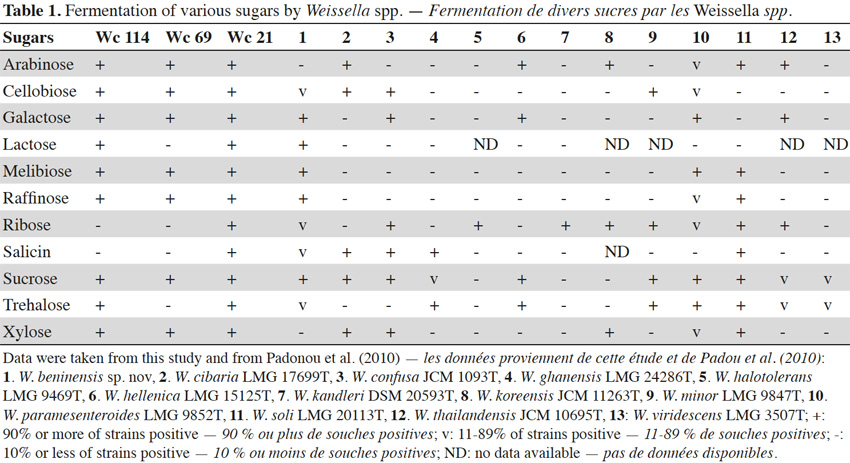
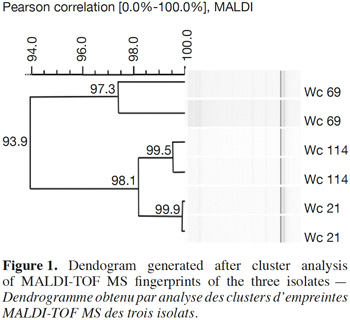
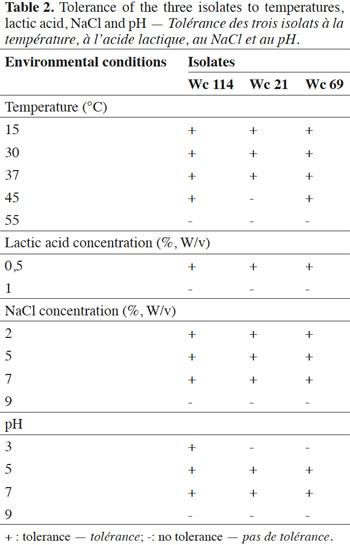
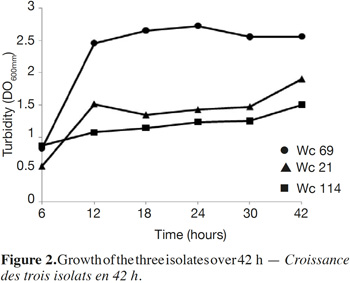
7In total, 111 Gram positive, catalase and oxidase negative rod strains (bacilli) were isolated from traditional cassava starters used for attiéké production. Ninety two of these strains (82.9%) were homofermentative and the remaining 19 ones (17.1%) were heterofermentative. It was observed that three heterofermentative isolates, named Wc 114 (isolated on M17), Wc 21 (isolated on BEA) and Wc 69 (isolated on MRS) present unusual non-motile short rod-sharped features with important lactic fermentation potentiality. On API 50 CHL kit, the three isolates show a similar fermentation profile except for ribose, salicin, D-trehalose and D-turanose (Table 1). The three strains were identified by MALDI-TOF MS technique as either Weissella cibaria or Weissella confusa (both species have similar MALDI-TOF MS profiles). Statistically, high values of similarity (Pearson correlation) > 99% were observed i.e. a Pearson correlation of 99.5% for isolates Wc 21 and Wc 114 and 99.7% for Wc 69 (Figure 1). The resulting dendrogram indicated that both isolates Wc 21 and Wc 114 were close to each other while they were relatively remote from isolate Wc 69. The three isolates were facultative anaerobic, with growth ability at 15, 30 and 37 °C. None of these isolates tolerated 55 °C but isolates Wc 114 and Wc 69 supported 45 °C, when Wc 21 did not. The three isolates grew at the initial concentrations of 0.5% and 7% for lactic acid and NaCl, respectively; but they were unable to grow at 1% lactic acid and 9% NaCl (Table 2). Only isolate Wc 114 grew at pH = 3. On MRS broth, at 37 °C, isolate Wc 114 had a higher growth followed by isolates Wc 21 and Wc 69 (Figure 2).Their growth started by the exponential phase (6-12 h) and then, the stationary phase occurred (12-30 h) followed by the decline phase (between 30-42 h). The microbial growth was accompanied by a rapid decrease of glucose content in the early hours of fermentation (6-18 h) and then stabilization appeared (Figure 3).
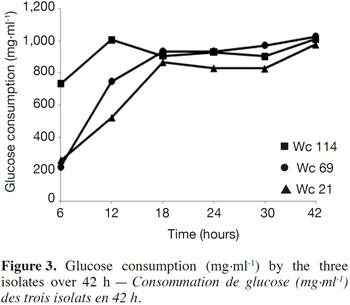
3.2. Technological properties of isolates
8The three isolates synthesized α-amylase and cell wall-degrading enzyme pectinase for isolate Wc 114, and they fermented raffinose which is an indigestible sugar. None of the strains synthesized β-glucosidase. In general, these isolates induced a rapid and significant decrease in pH of their growth medium accompanied by production of lactic acid (Figure 4). Indeed, all the three isolates induced a pH drop of MRS broth below 5.3 after 6 h and below 4.5 after 24 h. Isolates Wc 21 and Wc 114 induced a drop of the MRS broth pH below 4 after 12 h. Within the same period, the pH of MRS broth inoculated with isolate Wc 69 dropped to 4.30. At 42 h of fermentation, the final pH was 4.1, 3.7 and 3.5 for isolates Wc 69, Wc 21 and Wc 114, respectively. The yield of produced lactic acid was lowest for isolate Wc 69 compared to isolates Wc 114 and Wc 21. Total titratable acidity (expressed as percentage of lactic acid) increased significantly from 6 to 12 h. At the end of fermentation, lactic acid levels reached 1.63%, 1.36% and 0.73% for isolates Wc 114, Wc 21 and Wc 69, respectively.
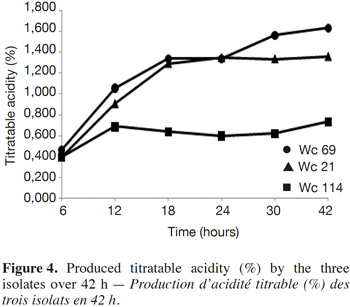
4. Discussion
9From former investigations, Lactobacillus is known as the dominant bacterial genus throughout cassava fermentation (Kostinek et al., 2007). However, this study showed that isolates Wc 114, Wc 21 and Wc 69 were heterofermentative bacilli, but the API 50 CHL kit was ineffective for their identification. Similar observation has been reported by Björkroth et al. (2002) concerning identification of strains of Weissella confusa and Weissella cibaria, respectively. The use of MALDI-TOF MS confirmed them as Weissella strains. Moreover, their fermentation profile was different from those usually reported for Weissella strains by Padonou et al. (2010a). Similar situation occurred for four species of Weissella named Weissella beninensis sp. nov. with unusual phenotypic and genotypic properties isolated from submerged fermenting cassava on MRS agar (Padonou et al., 2010b). In addition, Weissella ghanensis sp. nov. has recently been isolated from a Ghanaian cocoa fermentation (De Bruyne et al., 2008). By the MALDI-TOF MS, Wc 114 and Wc 21 have more similarities in their protein profiles and substantially similar properties. It is worth noting that MALDI-TOF MS is a rapid identification method that has been recently introduced in bacterial taxonomy (De Bruyne et al., 2011); it is a performing tool for LAB identification. Indeed, this technology identifies microorganisms via the generation of fingerprints of highly abundant proteins followed by correlation to reference spectra in a database. MALDI-TOF MS has been successfully applied to a number of taxa of Listeria species (Barbuddhe et al., 2008), Campylobacter spp. (Fagerquist et al., 2007; Grosse-Herrenthey et al., 2008), Streptococcus pyogenes (Moura et al., 2008), the Burkholderia cepacia complex (Vanlaere et al., 2006), Arthrobacter (Vargha et al., 2006), Leuconostoc spp., Fructobacillus spp., and Lactococcus spp. (De Bruyne et al., 2011), Lactobacillus spp., Weissella spp., Lactococcus spp., Pediococcus spp. (Doan et al., 2012). Although the profiles of the studied isolates suggest that they might belong to the taxons Weissella cibaria or Weissella confusa (percentage of identification exceeding 99%), further study is required to resolve their identity. More precisely, it would be interesting to characterize Wc 114, Wc 21 and Wc 69 using molecular methods which have a greater discriminatory power. Considering environmental conditions, many LAB are known to be acidophilic and so, would be tolerant to low values of pH. The three isolates presented an osmotolerant property (growth at 7% NaCl) and the isolates Wc 114 and Wc 69 were also thermotolerant, with abilities of growth and acidification up to 45 °C. These two properties suggest they could be suitable for industrial applications, especially for the thermotolerance in tropical regions where ambient temperature is generally higher than 30 °C. Indeed, this property makes it possible to reduce energy for cooling during production in bioreactor. Moreover, a high fermentation temperature also reduces contamination by other microorganisms. During industrial fermentation, as lactic acid is being produced by cells, alkali would be pumped into the broth to prevent excessive pH reduction. Hence, the free organic acids would be converted into their salts, resulting in the increase of osmotic pressure on cells. Therefore, high osmotolerance ability is a desirable feature for industrial LAB. Our results are in agreement with a statement of Collins et al. (1993) that some Weissella confuse strains can grow at 45 °C. Amylase and β-glucosidase activities are required for microbial starters to enhance digestibility and to decrease toxicity of cassava-derived foods (Kostinek et al., 2007; Vinodh et al., 2011). Also, pectinolytic enzyme produced by isolate Wc 114 is necessary for cassava root softening (Padonou et al., 2010a) although this interesting property was not required as a feature for the selection of LAB as starter culture for cassava dough fermentation into gari (Kostinek et al., 2007). Another interesting property of the studied strains was their ability to ferment raffinose, an indigestible sugar. In humans, these sugars are metabolized by microorganisms in the large intestine, liberating huge amounts of gas, which can then cause gastrointestinal disorders (LeBlanc et al., 2004). A further important feature for potential microbial starter strains is the ability to acidify its environment. It is worth recalling here that the acid production and the accompanying pH decrease are well-known to extend the lag phase of sensitive organisms including foodborne pathogens. The three LAB strains were considered as fast acid producers compared to studies of Kostinek et al. (2007) who have demonstrated that this potentiality is mainly found among Lactobacillus plantarum (94% strains isolated from fermenting cassava during gari production in Benin). Also, from gari fermentation process, the potential microbial starters induced a drop of the MRS broth pH to less than 3.9 after 48 h growth (Kostinek et al., 2007). A pH value below 3.9 constitutes a major food safety factor. Indeed, spoilage bacteria, as well as pathogens, notably those including members of Enterobacteriaceae, do not grow below this pH (Kostinek et al., 2007). The presence of coliform was mentioned in traditional cassava starters and in the cassava dough during attiéké production. Their presence would suggest possible fecal contamination, maybe through water or manufacturing materials or from the environment (Tetchi et al., 2012). In cassava dough used for attiéké production, the titratable acidity increases up to 0.6 ± 0.05% after 24 h (Coulin et al., 2006). Also, in traditional cassava starters, the titratable acidity ranges from 0.02-0.09% (Koussemon et al., 2010; Tetchi et al., 2012). Isolates enabling to reach a total titratable acidity > 0.7% are in principle suitable as starters for lactic acid fermentation of cassava dough. Kostinek et al. (2007) demonstrated that strains of Weissella could be selected as starter cultures for cassava dough fermentation. Thus, on the basis of their biochemical properties, two strains of Weissella paramesenteroides were selected as microbial starters for gari (a cassava fermented partially gelatinized granular product) production. In other works, the use of Weissella strains as starter cultures in food production was reported. The mechanical properties of pizza crusts were evaluated before and after their inoculation with a starter of microorganisms for which proteolytic activities are present (Lactobacillus sakei T56, Weissella paramesenteroides A51, Candida krusei G271) or absent (Lactobacillus sakei T58, Weissella paramesenteroides A58, Saccharomyces cerevisiae T22) (Pepe et al., 2003). Better viscoelasticity and better firmness were obtained with the proteolytic starter. Strains of Weissella cibaria, producing exopolysaccharides were used in monoculture or in mixed cultures as starters in the production of bread (Schwab et al., 2008; Zotta et al., 2008). All these findings show the fermentative abilities of Weissella strains and justify their choice as starter cultures for food production, particularly for cassava fermentation in tropical countries. However, before any industrial application it would be interesting to verify their potentialities for attieké production at small scale level.
5. Conclusions
10To the best of our knowledge, Weissella strains have never been reported among the fermenting microorganisms from traditional cassava starters used for attiéké production. The three isolates obtained in this study, identified as Weissella strains, present suitable technological properties and could be further exploited for industrial application for cassava dough fermentation. However, it would be interesting to test them as pure microorganisms at small-scale level during attiéké production.
11Acknowledgements
12We thank the WAAP (West Africa Agricultural Productivity Program) and AUF (Agence universitaire de la francophonie: University Agency of the Francophonie) for their financial support. We are also grateful to producers who had freely accepted to participate to this study and the Laboratory of Microbiology of University of Ghent from Belgium for the MALDI-TOF MS analysis.
Bibliographie
Akely P.M.T., 2012. Influence de la fermentation contrôlée, du pressage et de la granulation mécanisés du manioc (Manihot esculenta Crantz) râpé sur les caractéristiques physicochimiques et sensorielles de l’attiéké. PhD thesis: Nangui Abrogoua University, Abidjan (Côte d’Ivoire).
Barbuddhe S.B. et al., 2008. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol., 74, 5402-5407.
Björkroth K.J. et al., 2002. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int. J. Syst. Evol. Microbiol., 52, 141-148.
Collins M.D., Samelis J., Metaxopoulos J. & Wallbanks S., 1993. Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol., 75(6), 595-603.
Coulin P. et al., 2006. Characterization of the microflora of attiéké, a fermented cassava product, during traditional small-scale preparation. Int. J. Food Microbiol., 106(2), 131-136.
De Bruyne K. et al., 2008. Weissella ghanensis sp. nov. isolated from a Ghanaian cocoa fermentation. Int. J. Syst. Evol. Microbiol., 58, 2721-2725.
De Bruyne K. et al., 2011. Bacterial species identification from MALDI-TOF mass spectra through data analysis and machine learning. Syst. Appl. Microbiol., 34, 20-29.
Diallo Y. et al., 2013. Importance nutritionnelle du manioc et perspectives pour l’alimentation de base au Sénégal. Biotechnol. Agron. Soc. Environ., 17(4), 634-643.
Dicks L.M.T. & van Vuuren H.J.J., 1987. A modification of the hot-tube method for the detection of carbon dioxide produced by heterofermentative Lactobacillus strains. J. Microbiol. Methods, 6, 273-275.
Doan N.T. et al., 2012. Validation of MALDI-TOF MS for rapid classification and identification of lactic acid bacteria, with a focus on isolates from traditional fermented foods in Northern Vietnam. Lett. Appl. Microbiol., 55(4), 265-273.
Fagerquist C.K., Yee E. & Miller W.G., 2007. Composite sequence proteomic analysis of protein biomarkers of Campylobacter coli, C. lari and C. concisus for bacterial identification. Analyst, 132(10), 1010-1023.
Freiwald A. & Sauer S., 2009. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc., 4, 732-742.
Grosse-Herrenthey A. et al., 2008. Challenging the problem of clostridial identification with matrix-assisted laser desorption and ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe, 14(4), 242-249.
Holzapfel W.H., 2002. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol., 75(3), 197-212.
Kostinek M. et al., 2007. Characterisation and biochemical properties of predominant lactic acid bacteria from fermenting cassava for selection as starter cultures. Int. J. Food Microbiol., 114(3), 342-351.
Kouamé A.K., Djéni T.N., N’guessan F.K. & Dje M.K., 2012. Postprocessing microflora of commercial attieke (a fermented cassava product) produced in the south of Côte d’Ivoire. Lett. Appl. Microbiol., 56, 44-50.
Koussemon M.C. et al., 2010. Biochemical and microbial changes during traditional spontaneous lactic acid fermentation process using two varieties of cassava for production of a “Alladjan” starter. Int. Food Res. J., 17, 563-573.
LeBlanc J.G., Garro M.S. & Savoy de G.G., 2004. Effect of pH on Lactobacillus fermentum growth, raffinose removal, alpha-galactosidase activity and fermentation products. Appl. Microbiol. Biotechnol., 65(1), 119-123.
Moura H. et al., 2008. MALDI-TOF mass spectrometry as a tool for differentiation of invasive and non invasive Streptococcus pyogenes isolates. FEMS Immunol. Med. Microbiol., 53(3), 333-342.
Obilie E.M., Tano-Debrah K. & Amoa-Awua W.K., 2003. Microbial modification of the texture of grated cassava during fermentation into akyeke. Int. J. Food Microbiol., 89, 275-280.
Padonou S.W. et al., 2010a. Development of starter culture for improved processing of Lafun, an African fermented cassava food product. J. Appl. Microbiol., 109(4), 1402-1410.
Padonou S.W. et al., 2010b. Weissella beninensis sp. nov., a motile lactic acid bacterium from submerged cassava fermentations, and emended description of the genus Weissella. Int. J. Syst. Evol. Microbiol., 60, 2193-2198.
Pepe O. et al., 2003. Effect of proteolytic starter cultures as leavening agents of pizza dough. Int. J. Food Microbiol., 84, 319-326.
Schwab C., Mastrangelo M., Corsetti A. & Ganzle M., 2008. Formation of oligosaccharides and polysaccharides by Lactobacillus reuteri LTH5448 and Weissella cibaria 10M in sorghum sourdoughs. Cereal Chem., 85, 679-684.
Tetchi F.A., Oluwa W.S., Kakou A.C. & Amani N.G., 2012. Effect of cassava variety and fermentation time on biochemical and microbiological characteristics of raw artisanal starter for attiéké production. Innovative Rom. Food Biotechnol., 10, 40-47.
Vanlaere E. et al., 2006. Identification of Burkholderia cepacia complex using MALDI-TOF mass spectrometry. J. Cystic Fibrosis, 5, S34.
Vargha M., Takats Z., Konopka A. & Nakatsu C.H., 2006. Optimization of MALDI-TOF MS for strain level differentiation of Arthrobacter isolates. J. Microbiol. Methods, 66, 399-409.
Vinodh A.E. et al., 2011. Biomass production and small-scale testing of freeze-dried lactic acid bacteria starter strains for cassava fermentations. Food Control, 22, 1-7.
Zotta T. et al., 2008. Characterization of lactic acid bacteria isolated from sourdoughs for Corsetto, a traditionnal bread produced in Basilicata. World J. Microbiol. Biotechnol., 24, 1785-1795.
