- Home
- Volume 24 (2020)
- Numéro 1
- Genetic characterization of promising high-yielding cashew (Anacardium occidentale L.) cultivars from Côte d'Ivoire
View(s): 0 (0 ULiège)
Download(s): 0 (0 ULiège)
Genetic characterization of promising high-yielding cashew (Anacardium occidentale L.) cultivars from Côte d'Ivoire

Editor's Notes
Received 6 August 2018, accepted 27 November 2019, available online 21 January 2020
This article is distributed under the terms and conditions of the CC-BY License (http://creativecommons.org/licenses/by/4.0)
Résumé
Caractérisation moléculaire des cultivars d’anacardiers potentiellement hauts producteurs en Côte d’Ivoire
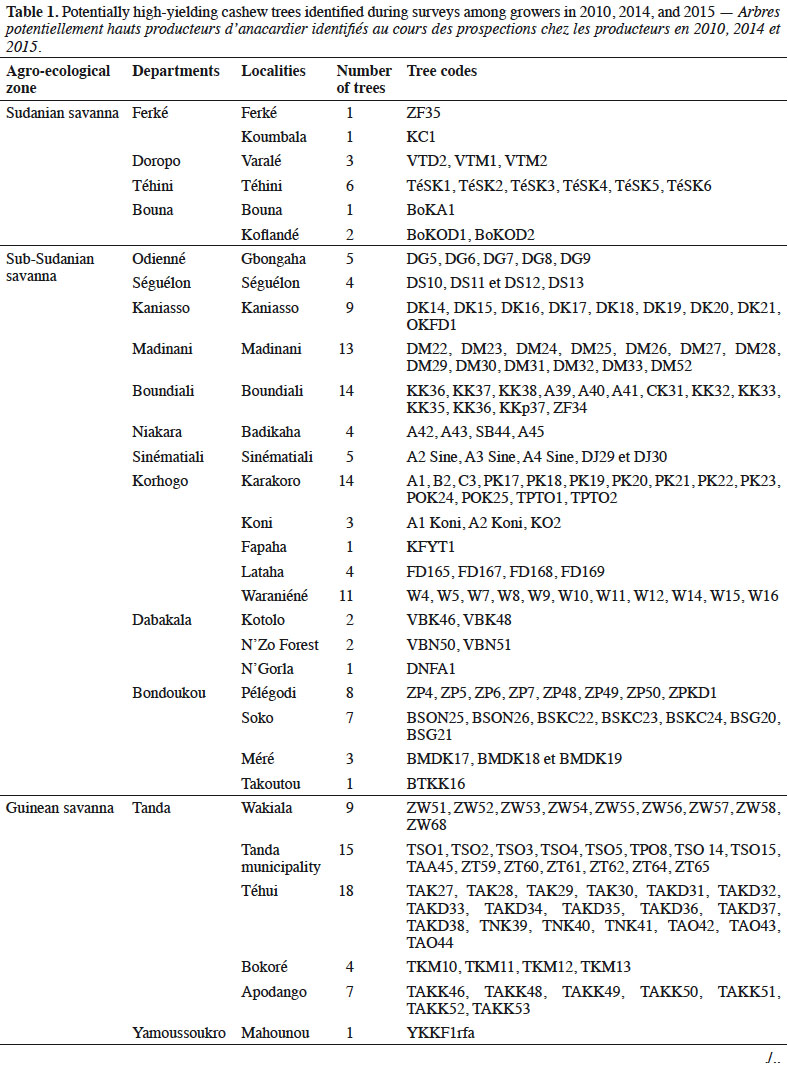
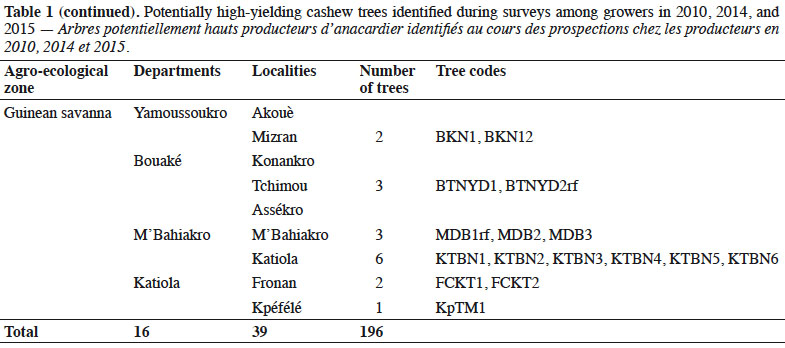
Description du sujet. L'anacardier a été introduit en Côte d'Ivoire en 1951 à des fins écologiques. De 1972 à 1980, il y a eu une transformation des plantations forestières en vergers et l'introduction de la variété ‘ Jumbo ’ du Brésil. Cela a été suivi d'un mélange de cette variété avec les populations d'arbres préexistantes. L'expédition de germoplasmes, réalisée en 2010 et de 2014 à 2015 dans toutes les principales zones de production de noix de cajou en Côte d'Ivoire, a aidé à identifier 209 cultivars à haut rendement. Bien que les caractéristiques morphologiques de ces cultivars aient été évaluées, la diversité et la structure génétiques des germoplasmes ne sont que peu connues.
Objectifs. L'objectif de cette étude est d'évaluer la diversité génétique des cultivars d’anacardier à haut rendement pour une meilleure utilisation dans les programmes de sélection.
Méthode. Une extraction de l'ADN a été effectuée à l'aide de Qiagen DNeasy Plant Mini Kits. Les analyses PCR ont été effectuées avec 18 marqueurs SSR.
Résultats. Les résultats ont révélé deux populations d’anacardier initialement introduites en Côte d’Ivoire. La richesse allélique obtenue a été de 3,56 ± 1,45 allèles en moyenne par locus. La valeur FIS indique un déficit global d'hétérozygotie de 0,332 ± 0,076. La différenciation moyenne de la population est de FST = 0,014 ± 0,004. Une grande partie de la variabilité génétique totale se trouve au niveau intra-population (98,6 %), contre seulement 1,4 % de cette variabilité attribuée aux différences entre les populations. Le flux moyen des gènes est de 22,528.
Conclusions. L'important flux génétique maintient une forte diversité génétique intra-population. Cela reflète une variabilité génétique exploitable à long terme dans le choix et la conservation.
Abstract
Description of the subject. Cashew was introduced to Côte d'Ivoire in 1951 to control erosion and reforest cutover lands. From 1972 to 1980, natural forest plantations were converted to fruit orchards and were supplemented by the ‘Jumbo’ cashew variety imported from Brazil. Germplasm expeditions conducted in 2010 and 2014–2015 identified 209 high-yielding cultivars in the major cashew growing areas of Côte d’Ivoire. Although the morphological characteristics of these cultivars have been assessed, little is known about the genetic diversity and genetic structure of the germplasm collection.
Objectives. The objective of the study was to evaluate the genetic diversity of high-yielding cashew cultivars for better use in breeding programs.
Method. We performed DNA isolation using Qiagen DNeasy Plant Mini Kits and PCR analysis with 18 SSR markers.
Results. We identified the first two introduced populations of cashew in Côte d’Ivoire. The average allelic richness is 3.56 (± 1.45) alleles per locus, the fixation index (FIS) indicates an overall heterozygosity deficit of 0.332 (± 0.076), and the average population differentiation (FST) is 0.014 (± 0.004). Much of the total genetic variability occurs at the intra-population level (98.6%), compared to only 1.4% variability attributable to differences between populations. Average value of gene flow is 22.528.
Conclusions. Gene flow within cashew populations maintains high intra-population genetic diversity. This flow rate reflects a long-term exploitable genetic variability for use in selection and conservation.
Table of content
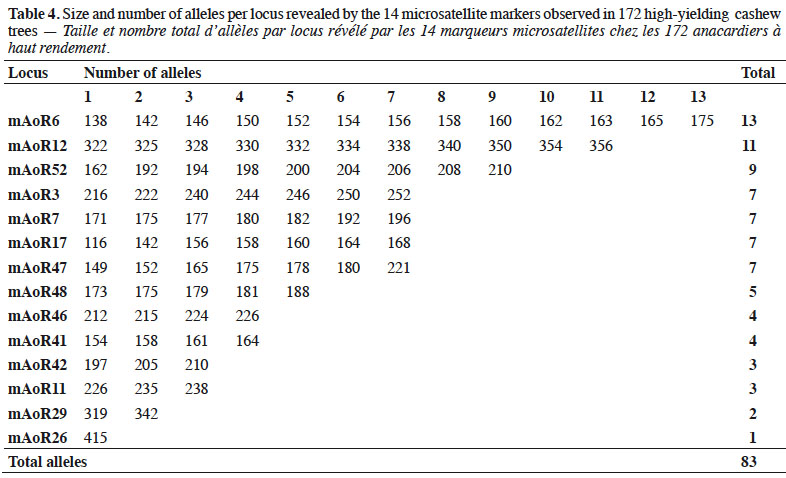
1. Introduction
1Cashew (Anacardium occidentale L., Anacardiaceae) is a tree species native to Brazil (Trevian et al., 2005). Cashew was first introduced in Côte d'Ivoire in 1951 to reforest cutover lands and inhibit soil erosion (Goujon et al., 1973). Between 1959 and 1960, cashew forest plantation programs were implemented by seeding nuts in the northern and center parts of Côte d'Ivoire and then throughout the entire Sudano-Guinean savanna zone (Goujon et al., 1973). The planting of cashew in initial forest restoration projects was also economically beneficial to the region. Subsequently, the former Fruit and Vegetable Development Corporation (SODEFEL) introduced the ‘Jumbo’ variety to the region, which they planted in two separate blocks in Badikaha, a small town near the city of Ferkessédougou. Half-sib progenies of the ‘Jumbo’ variety were used to establish the first cashew germplasm with 234 trees at the National Center for Agronomic Research (CNRA) Station in Lataha. A germplasm collection expedition, which was conducted by the research team of the CNRA from 2010 to 2015, identified 209 high-yielding trees (HYTs).
2Since 2015, Côte d’Ivoire has become the world’s largest producer of cashew nuts, producing 700,000 tons (Cashew Info, 2016). However, Ivorian cashew orchards are largely composed by non-selected plant material and mostly characterized by inefficiently planted trees, which together contribute to relatively low nut yields (448 kg·ha-1 on average) (Cashew Info, 2014), compared to the minimum of 1,000 kg·ha-1 harvested from orchards in India and Brazil. A recent increase in cashew production in Côte d'Ivoire is due in large part to the expansion of land under cultivation, which replaced natural vegetation and areas previously devoted to other crops. The land area under cashew tree cultivation was about 234,000 ha in 2002, but is estimated in 2016 at about 1,567,000 ha (a six-fold increase).
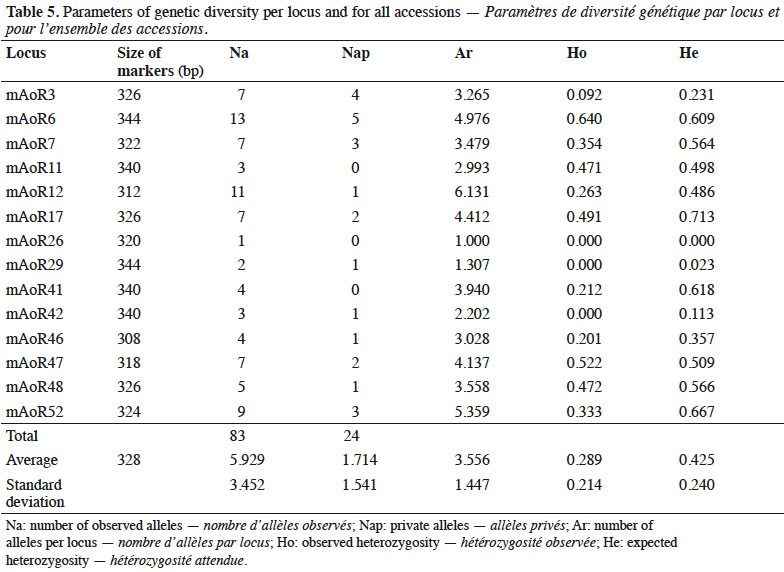
3One approach for increasing cashew tree yield and improving the quality of nuts grown in Côte d'Ivoire is to select genotypes with traits of interest from existing planting material and to use them in breeding program to develop superior planting (Aliyu & Awopetu, 2007). High-yielding trees identified in orchards and trees residing in the CNRA germplasm together provide genetic resources that can be exploited to genetically improve cashew orchards in Côte d'Ivoire. The material hold in Lataha collection has already been agro-morphologically characterized, which has defined various groups of accessions by their phenotypic traits (Djaha et al., 2014; Kouakou et al., 2018). The judicious exploitation of the identified traits will enable the selection and creation of new cashew varieties in Côte d'Ivoire. However, because little is known about the genetic diversity and genetic structure of this collected plant material, it is essential to determine its genetic diversity before seeds from the collection can be used for the genetic improvement of varieties.
4Selection of cashew varieties is usually obtained using traditional morphometric methods, which identify interesting and potentially useful phenotypic characteristics, such as sizes and mass of nuts, sizes of whole fruits, colors of apples, sex ratios, sizes of plants, the lengths of panicles, and yields of trees (Chabi Sika et al., 2013). Although phenotypic characteristics can provide useful metrics, their usefulness for genetic selection is often diminished by environmental effects on growth (Aliyu & Awopetu, 2007). To circumvent the effects of environment on morphology, molecular markers, which are stable and unaffected by the local environmental conditions, are more appropriate for determining genetic variability, identifying varieties, and managing genetic resources (Adoukonou-Sagbadja et al., 2007). Thus, molecular markers should be used to assess the genetic diversity of high-yielding cashew trees in Côte d'Ivoire, which in turn could be used to better utilize local germplasm for commercial benefit.
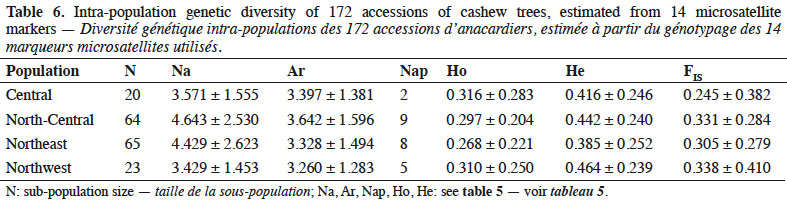
5Molecular markers used to study genetic diversity in cashew tree include data on microsatellites (SSRs) (Croxford et al., 2006). SSRs provide robust and efficient markers because they are highly polymorphic, co-dominant, multi-allelic, and highly reproducible in nature (Williams et al., 1990). For these reasons, this study used microsatellite markers to assess the genetic diversity of high-yielding cashew trees identified in Côte d'Ivoire.
2. Materials and methods
2.1. Plant material
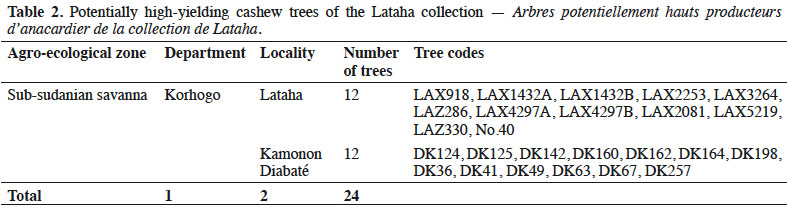
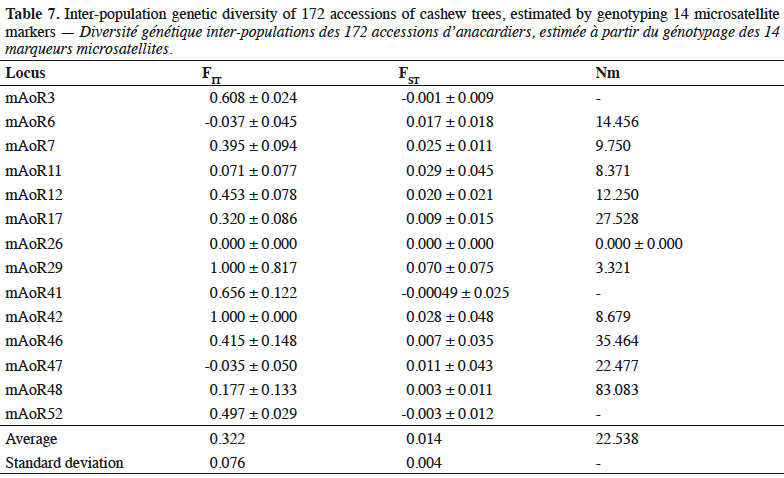
6We collected germplasm from cashews in commercial orchards throughout the major cashew-producing regions of Côte d'Ivoire during the 2010, 2014, and 2015 peak vegetative growth seasons (i.e., June–July) (Figure 1). Cashews growers were asked to identify specific trees that in past years had consistently produced high yields (growers were not asked to quantify production). Based on grower’s qualitative evaluations, we selected 221 trees to identify their SSRs (Tables 1 and 2). We obtained an average of 25 scions (young shoots) per tree and shipped them to the nearest cashew research station (CNRA site).
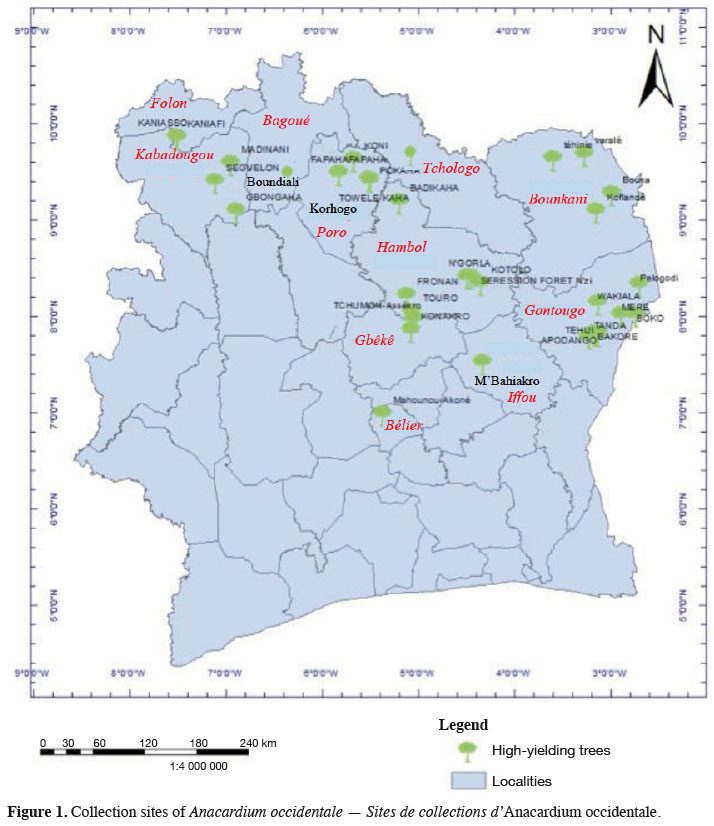
2.2. Methods
7Grafting. At the experimental stations, we top-grafted collected scions onto 45-day-old seedlings previously prepared as rootstock. We segregated grafted plants by geographic origin and maintained the plants in a nursery for 40 days to produce young leaves.
8DNA isolation. We isolated DNA from each nursery plant from 100 mg of fresh leaves, collected from 221 leaf samples from the three CNRA sites (Lataha, Madinani, and Tanda). We cleaned each sampled leaf, froze it in liquid nitrogen, and ground it. We then extracted DNA using the Qiagen Kit method (Dneasy Plant Mini Kit/www.qiagen.com, 2015) according to the manufacture’s instruction, but with modification at the grinding stage. Leaves were ground in liquid nitrogen to facilitate cell lysis. We added an extraction buffer (1.5 ml) while grinding. We removed debris and other chemical and biochemical compounds (proteins, mineral salts, and lipids) by centrifugation. We precipitated out DNA by adding alcohol purified using the Qiagen kit column and diluted it into nuclease-free water. We quantified the DNA using a Nanodrop UV-vis 2000 spectrophotometer (Thermo scientific, USA) and stored the sample in a freezer maintained at -30 °C.
9Microsatellite markers. We performed in vitro amplification using the PCR (Polymerase Chain Reaction) method with 18 microsatellite primers (Croxford et al., 2006). We used each primer to amplify 2 ng of DNA in 10 μl of reaction mixture using the GeneAmp instrument (Applied Biosystem). The characteristics of these primers are presented in table 3.
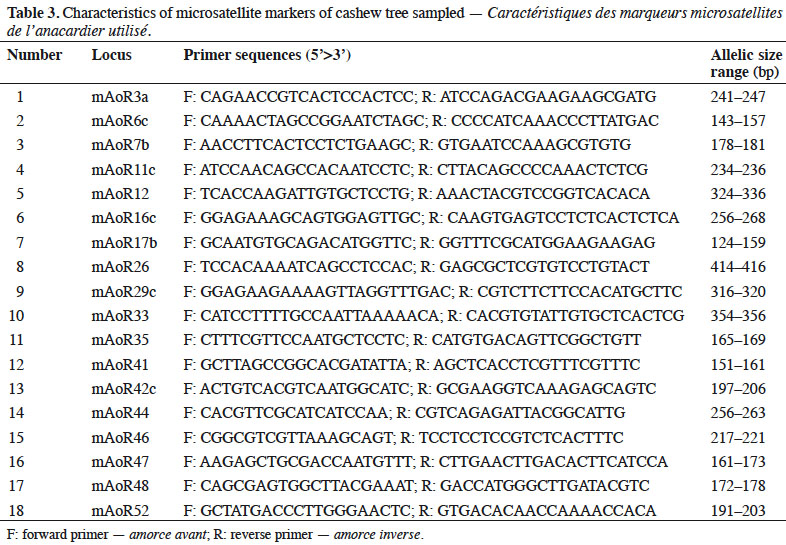
10Data analysis
11Analysis of PCR products. We initially scored electrophoregram profiles based on fragment size, but only unambiguous amplicons were scored. We performed the scoring of microsatellite amplicons with the SAGA GT TM software program (LI-COR, Inc. Lincoln, Nebraska, USA) and then determined each genotype after several re-readings, paying particular attention to the presence of artifacts in the profile.
12Assessment of genetic diversity parameters. At the intra-population level, we estimated genetic diversity using the following assessment parameters:
13– allelic richness (Ar), obtained with Fstat software (F-STAT 2.9.3.2: Institute of Ecology, Lausanne, Switzerland);
14– Nei’s genetic diversity (He), equivalent to expected heterozygosity under the Hardy-Weinberg hypothesis (Nei, 1978), obtained with Popgen 32 software;
15– proportion of heterozygous loci observed per individual (Ho), obtained with Popgen 32 software;
16– fixation index (FIS) (i.e., inbreeding coefficient).
17Assessment of inter-population genetic diversity. At the inter-population level, we estimated genetic diversity using the following assessment parameters:
18– total genetic diversity of all the populations considered as a single population (FST), determined using the Fstat software;
19– fixation index of an individual of the population in relation to the total (FIT), determined using the Fstat software;
20– estimated gene flow (Nm) from FST, where Nm = 0.25 (1–Fst) Fst-1;
21– analysis of molecular variance (AMOVA). AMOVA is a method for directly estimating the degree of differentiation between populations (genetic diversity between regions) on the basis of molecular data. We used Harlequin software version 3.1 to perform the AMOVA at the p < 0.05 level of significance.
22Genetic distance and dendrogram construction. Another approach for studying genetic differentiation between individuals or populations is to analyze the degree of similarity between them. A distance matrix can be constructed when more than two populations are analyzed and all possible pairs of distances are estimated. From such a distance matrix, a multivariate analysis can be performed to describe the structure of the genotype on scatter plot. The groups can then be represented by a dendrogram (i.e., a tree diagram expressing kinship relationships between accessions of cashew trees). In our study, we used similarity indices to analyze individual’s proximity. These indices were expressed as genetic distances, wherein the genetic distance for each pair was equal to 1.0 minus its similarity index. Thus, all distance values ranged between zero and one. A phylogenetic tree diagram (dendrogram) could then be generated with the neighbor-joining method. This method is the best for identifying the most genetically similar individuals, consequently we used it to calculate similarity indices (Bennett et al., 1997).
23We used the matrix of estimated distances between individuals in the total population to construct dendrograms. A zero distance between two individuals suggested identical relative to the compared loci. In contrast, wider distances reflected more divergence between compared individuals (Ould Ahmed et al., 2010). We used Darwin 6.0.11 software to construct the dendrogram to visualize relationship.
3. Results
3.1. Polymorphism of microsatellite markers
24We analyzed 221 individuals with the 18 microsatellites (Table 3). Of these 18 SSRs, those with more than 10% missing data (n = 4) were excluded from further analysis. Similarly, we also excluded individual leaf samples with more than 50% missing data. The obtained genotype information was composed of 14 markers for 172 individuals.
25Our paired comparisons revealed that 14 loci did not show a general signal of linkage disequilibrium, suggesting that these loci could be considered independently in subsequent analyses. The 14 microsatellite markers we finally used generated 83 alleles characterized by fragment sizes ranging from 116 to 415 base pairs (bp) (Table 4). Thirteen SSR markers of the 14 give polymorphic profiles which revealed 2–13 alleles. The most informative markers revealed 7–13 alleles within populations.
3.2. Genetic variability
26Expression of genetic diversity among the population of cashew
27Allelic richness. We found an average of 3.56 (± 1.45) alleles per locus across all accessions (Table 5). We detected 24 private alleles within the cashew tree accessions (Table 5). These private alleles constitute alleles found exclusively in particular accessions, representing 29% (24/83) of the alleles in all the cashew accessions we found.
28Heterozygosity and allelic richness. We determined that expected heterozygosity was 0.425 ± 0.240, whereas observed heterozygosity was 0.289 ± 0.214, showing a deficiency of heterozygotes (Table 5).
29We segregated cashew accessions into four populations based on the regions from which we obtained them (central, north-central, northeast, and northwest). Based on our analysis of the allelic richness of the various accessions from the four regions, we determined that the accessions of plants from the central region possessed allelic richness (number of alleles per locus) that was similar to the one of the other regions (Table 6). However, of the 24 private alleles (Nap) we identified, the highest percentage (37.5%) occurred in cashew plants collected from the north-central region. In contrast, the lowest percentage (8.3%) of Naps occurred in plants collected from the central region. The north-central and northeast were very similar in Naps, they co-dominated (Table 6).
30Fixation index (inbreeding coefficient). The fixation index is a measure of the deficiency or excess of heterozygote through the ration of individuals found in the heterozygous state (Ho) and the expected heterozygote (He). All four populations showed positive fixation indices (FIS) (Table 6), among which cashew trees from the central region exhibiting the lowest value (FIS = 0.245 ± 0.382).
31Expression of inter-population genetic diversity and gene flow
32FIT Parameters, Wright’s F statistics (fixation index FST), and gene flow. Table 7 shows the fixation index (FIT) (which quantifies degree of genetic differentiation of an individual of the population relative to the total population) and the FST index (which quantifies the degree of genetic differentiation between populations). The FIT value indicated that there is a 33% deficit of heterozygotes (0.332 ± 0.076) when taking into account the four populations examined as single population. The average population differentiation was Fst = 0.014 ± 0.004, which is considered to be low. This means that a large proportion (98.6%) of the total genetic variability in cashews of Côte d’Ivoire can be explained by the intra-population variation and that 1.4% of this variability is attributed to the differences between populations of the cashew accessions we studied. The average gene flow (Nm) was 22.528 (Table 7).
33Dendrograms. The hierarchical analysis we conducted using all accessions and a dendrogram showed that the two main groups (clusters) of cashews could be segregated into six sub-groups (Figure 2). The first cluster segregated into four groups, identified hereafter as subgroups A, B, C, and D, whereas the other cluster segregated into two subgroups (E and F). The number of accessions varied by subgroup (range 21–39), as follows: subgroup A (39 accessions), subgroup B (27 accessions), subgroup C (21 accessions), subgroup D (20 accessions), subgroup E (39 accessions), and subgroup F (26 accessions). Accessions that compose each of these groups originated from different geographical regions of Côte d’Ivoire. In each subgroup, we found duplicate individuals within HYTs which were genetically identical. All of these “duplicated” individuals were collected from the same orchards. For example, in subgroup A, the following HYT tree duplicates originated from the same location (Kaniasso, Odienné) and same orchard (trees DK14, DK15, and DK16). Likewise, the KTTB4 and KTTB6 accessions occurred in the same orchard in Katiola.
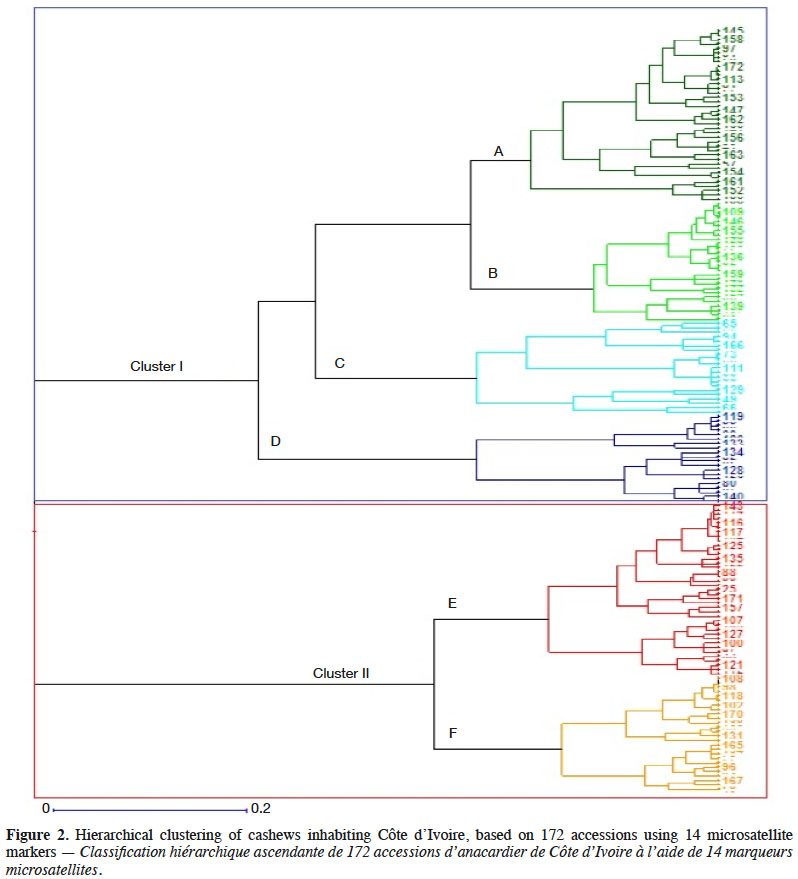
34Tree LAZ330 from Lataha, a genotype distributed among many growers, is genetically similar to all the aforementioned trees. The results of our molecular characterization revealed that the three trees disseminated by CNRA, trees LAX3264, LAX4297, and LAZ330, are different genotypes. Genotype LAX4297, which was initially considered a single tree, is actually comprised of two genetically distant trees. Tree LAX4297 B (red-colored apple) belongs to subgroup B, whereas LAX4297 A (yellow-colored apple) belongs to subgroup C.
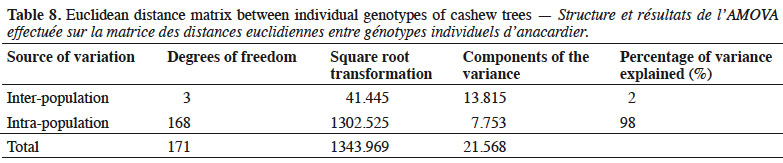
35Our analysis of molecular variance (AMOVA), which determined intra-population and inter-population genetic diversity based on the genetic distance matrix using individual’s genotype (Table 8), showed that only 2% of the total variation observed could be attributed to differences between populations. The majority of variation (98%) was due to differences within populations.
4. Discussion
36Our study used SSR markers to explore the genetic diversity of cashew accessions in Côte d’Ivoire. Among the SSR markers we used, 13 were polymorphic and so could serve as a reference for further cashew analyses in Côte d'Ivoire. These polymorphic markers could also be used to manage germplasm out-planting programs for long-term conservation and use.
37We found high allelic richness in all accessions from the various regions we studied because cashew is an outcrossing tree. The populations analyzed might reflect long-term genetic diversity that can be exploited in a breeding program to improve yield and nut quality. Moreover, identifying trees with high allelic richness is necessary for conserving cashew germplasm. Allelic richness data are also useful for managing germplasm collections and gene banks in terms of genetic diversity (Bataillon et al., 1996). The number of private alleles found within cashew accessions is important to know because these alleles show specific identity of genotypes and might explain specific characteristics of genotypes carrying these private alleles.
38Our results, showing that the allelic richness of accessions was similar for cashew plants collected from the four geographic regions we examined, suggest common origin of the material found in different region. Therefore, setting germplasm collection could be made using cashew germplasm from one region with additional accessions carrying private alleles in other regions. One implication of this insight into collection strategies is that for preserving accessions in gene banks, an identical number of high-yielding trees can be introduced while maintaining the same number of alleles per locus. This means that on the basis of allelic richness, the preservation of 20 accessions (the lowest number of accessions from the central region) per region is sufficient for maintaining the allelic richness of the entire germplasm.
39The number of private alleles in cashews of Côte d’Ivoire might reflect the sizes and locations of the first introduced populations. Thus, to preserve all specific alleles in the germplasm, it would be essential to maintain one copy of all duplicated accessions across geographic/climatic regions.
40Among the four cashew populations we collected (each associated with a specific geographic region), inbreeding coefficient was lowest in the central region, possibly because growers from this region could most easily exchange seeds with the other regions. The subsequent open crossing of introduced germplasms is probably responsible for the higher heterozygosity in the central region and its lower homozygosity than other regions. In contrast, north-central, northeast and northwest showed higher inbreeding coefficients. According to Ould ahmed et al. (2010), inbreeding (mating between an individual and its ascendants, collaterals, and/or descendants) modifies genotypic frequencies with a consequent loss of genetic variability over generations. Because the heterozygote levels are higher in the central region than in the other three regions, this region must have the highest relative diversity. Given that the central region is located near the other regions, its genetic richness might have benefited from having more seeds and more varied germplasm imported to it than the other regions.
41The difference between expected heterozygosity and observed heterozygosity might be due to evolutionary factors that occurred in the studied accessions or internal genetic factors (such as gene incompatibility). In addition, cashew grower’s preference when they selected seeds for establishing new orchards might be one of the reasons. Indeed, when establishing new cashew orchards, some producers used seeds from a single tree with good traits (preferentially high-yielding and large-nuts). Moreover, according to Ould Ahmed et al. (2010), the genetic makeup of a given population can vary over time in response to evolutionary forces that in turn affect the heterozygosity of the population relative to the Hardy-Weinberg equilibrium.
42Considering the four populations we studied, we regard the average population differentiation (FST) that we observed to be low (0.014 ± 0.004), indicating that the genetic diversity of Ivorian cashew trees could be possibly due to a common origin of cashew trees introduced to Côte d'Ivoire. This low genetic diversity might be due to the fact that the initial seeds used to establish orchards were probably not focused on yielding but resilience to dry environment and so were insufficient for guaranteeing a high diversity of plant material. Archak et al. (2009) found a similarly low genetic diversity among cashew trees across India and that most trees were introduced to India about 400 years ago. Masawe & Kapinga (2013) found that most other countries (except for Brazil) also do not have very rich cashew gene banks.
43About 23 migrants move from one accession to another within a generation. This reflects an important exchange of genes. This important gene flow might maintain the high intra-population genetic diversity we observed in Côte d'Ivoire.
44Our dendrogram showed two main genetically distinct groups in Côte d’Ivoire, from which we can infer that two distinct populations of cashew were introduced to Côte d’Ivoire. However, this result cannot indicate how cashew trees were introduced into the country. In addition, little is known about from where the subgroups of these two main groups were originate. Additional studies with samples from other regions of the world could provide insight into the evolution of the cashew.
45In each subgroup, we found genetically identical duplicated individuals (twins) of HYTs located in the same orchards. Such high levels of redundancy in accessions of cashew cultivars were also observed by Aliyu (2012). Some duplicated genotypes might be due to a limited number of markers used wherein all individuals identical at these loci are considered to be duplicates. Our results also reveal that all these propagated accessions originated from the same region. The accessions might have originated from the same orchard from which they were then disseminated to other orchards and regions. Our results suggest that cashew seeds introduced in Côte d'Ivoire were used to establish one orchard in Badikaha and another in Natiokobadara, and that a very large part of all orchards in Côte d'Ivoire might have been established using seeds from these two orchards. Archak et al. (2009) made similar observations and identified four distinct genetic groups. However, none of those four groups was restricted to a single region (i.e., all groups are almost equally represented in the four geographic populations). Thus, Archak et al. (2009) concluded that cashew orchard in Côte d'Ivoire were initially established at a single location with material introduced from a single area of India from which seeds were disseminated by humans to other areas of Côte d'Ivoire.
46The trees in our Lataha collection, which were released to growers, are progenies of trees from Badikaha and Natiokobadara (Djaha et al., 2014). The results of our molecular characterization revealed that three trees disseminated by the CNRA (trees LAX3264, LAX4297, and LAZ330) possess distinct genotypes. Genotype LAX4297, which we initially considered to have originated from a single tree, is actually comprised of two genetically distant trees, indicating that an off-type genotype was released under the same name. Trees LAX4297 B and LAX4297 A might have originated from two different nuts planted on the same hill or planting hole whose trunks eventually fused into a single tree. Architecturally, although these two trees now appear to be a single tree, it has two primary branches, one comprised of genotype LAX4297 A and the other comprised of genotype LAX4297 B. Genotype LAX4297 A was planted in the Ferké timber yard, whereas genotype LAX4297 B was planted in the Tanda timber yard. Genotypes LAX3264 and LAZ330 belong to subgroups C and A (respectively). Therefore, the CNRA currently distributes four genotypes to growers for establishing new orchards (rather than three).
5. Conclusions
47Genetic diversity exists among cashew orchards in Côte d'Ivoire and this diversity can be exploited to build a long-term program to select and preserve cashew germplasm. The difference we found between expected and observed heterozygosity might be due to the strategy that Ivorian cashew producers use in selecting trees for establishing new orchards. The trees across the four cashew-growing zones of Côte d'Ivoire showed outcrossing expressed by positive inbreeding coefficients (FIS), with a lower value for the central region than for the other three regions, expressing that the central region has the highest genetic diversity. Significant gene flows maintain this genetic diversity.
48The dendrogram of sample analyzed revealed two distinct populations of cashew trees inhabiting Côte d’Ivoire, comprised of six subpopulations. The grouping of HYT descendants performed without regard to geographic origins confirmed that cashew trees in all areas under cultivation have been disseminated from the northern part of Côte d’Ivoire.
Acknowledgements
49The authors warmly thank the Cotton and Cashew Council, which funded this study through the Interprofessional Fund for Agricultural Research and Council (FIRCA) with the West and Central African Council for Agricultural Research and Development (WECARD).
Bibliography
Bibliographie
Adoukonou-Sagbadja H. et al., 2007. Genetic diversity and population differentiation of traditional fonio millet (Digitaria spp.) landraces from different agro-ecological zones of West Africa. Theor. Appl. Genet., 115, 917-931, doi.org/10.1007/s00122-007-0618-x
Aliyu O.M. & Awopetu J.A., 2007. Assessment of genetic diversity in three populations of cashew (Anacardium occidentale L.) using protein-isozyme-electrophoretic analysis. Genet. Resour. Crop. Evol., 54, 1489-1497, doi.org/10.1007/s10722-006-9138-9
Aliyu O.M., 2012. Genetic diversity of Nigerian cashew germplasm. In: Mahmut Caliskan, ed. Genetic diversity in plants, 163-184, http://cdn.intechopen.com/pdfs/31477.pdf, (12/04/2019).
Archak S., Gaikwad A.B., Swamy K.R. & Karihaloo J.L., 2009. Genetic analysis and historical perspective of cashew (Anacardium occidentale L.) introduction into India. Genome, 52, 222-230, doi.org/10.1139/g08-119
Bataillon T.M., David J.L. & Schoen D.J., 1996. Neutral genetic markers and conservation genetics: simulated germplasm collections. Genetics, 144, 409-417.
Bennett L.B., Shriver M.D. & Bowcock A.M., 1997. Markers and methods for reconstructing modern human history. DNA Sequence, 8, 329-342.
Cashew Info, 2014. Cashew Handbook 2014 – Global Perspective. A product of www.cashewinfo.com. Bengaluru, India: Foretell Business Solutions Private Limited.
Cashew Info, 2016. Country profiles: Côte d’Ivoire, http://www.cashewinfo.com/country_profiles/Cote%20d'Ivoire.pdf, (10/07/2017).
Chabi Sika K. et al., 2013. Indigenous knowledge and traditional management of cashew (Anacardium occidentale L.) genetic resources in Benin. J. Exp. Biol. Agric. Sci., 1, 375-382.
Croxford A.E., Robson M. & Wilkinson M.J., 2006. Characterization and PCR multiplexing of polymorphic microsatellite loci in cashew (Anacardium occidentale L.) and their cross-species utilization. Mol. Ecol. Notes, 6, 249-251, doi.org/10.1111/j.1471-8286.2005.01208.x
Djaha A.J.B. et al., 2014. Diversité morphologique des accessions d’anacardier (Anacardium occidentale L.) introduits en Côte d’Ivoire. Rev. Ivoir. Sci. Technol., 23, 244-258.
Goujon P. et al., 1973. Études sur l’anacardier. Bois For. Trop., 151, 27-29,
Kouakou C.K. et al., 2018. Agro-morphological characterization of cashew (Anacardium occidentale L.) ecotypes from North-Central of Côte d’Ivoire. Fruits, 73(5), 255-264, DOI: 10.17660/th2018/73.5.1
Lebailly P., Lynn S. & Seri H., 2012. Étude pour la préparation d’une stratégie pour le développement de la filière anacarde en Côte d’Ivoire. Rapport Diagnostic, AGRER Consortium, 143 pages
Masawe P.A.L. & Kapinga F.A., 2013. Aspects of cashew hybridization by control hand pollination in Tanzania. In: Masawe P.A.L. et al., eds. Proceedings of the second international Cashew conference, Kampala, Uganda, 26-29 April 2010. Wallingford, UK: CAB International, 5-14.
Nei M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583-590.
Ould Ahmed M., Ben Salem F., Bedhiaf S. & M’Naouer D., 2010. Analyse moléculaire de la diversité génétique des dromadaires (Camelus dromedarius) en Tunisie. Biotechnol. Agron. Soc. Environ., 14, 399-408.
QIAGEN®, 2015. DNeasy® Plant Handbook.
Rey J.Y., 1998. La noix de cajou en Côte d'Ivoire en forte progression. Fruitrop, 51, 12-13.
Trevian M.T.S. et al., 2005. Characterisation of alkyl phenols in cashew (Anacardium occidentale L.) products and assay of their antioxidant capacity. Food Chem. Toxicol., 44, 188-197, https://doi.org/10.1016/j.fct.2005.06.012
Williams J.G. et al., 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl. Acids Res., 18, 6531-6535, doi.org/10.1093/nar/18.22.6531






