- Home
- Volume 28 (2024)
- Numéro 1
- A validated real-time PCR test for simultaneous detection of Gallus gallus and Meleagris gallopavo in feed instead of an impossible poultry test
View(s): 0 (0 ULiège)
Download(s): 0 (0 ULiège)
A validated real-time PCR test for simultaneous detection of Gallus gallus and Meleagris gallopavo in feed instead of an impossible poultry test

Résumé
Un test PCR en temps réel validé pour la détection simultanée de Gallus gallus et Meleagris gallopavo dans les aliments pour bétail au lieu d’un test volaille impossible
Description du sujet. L’utilisation de toutes protéines animales transformées (PATs) dans les aliments pour animaux a été interdite dans l’UE en raison de l’épidémie d’encéphalopathie spongiforme bovine (ESB). Cette interdiction totale a été progressivement levée à partir de 2013 pour les PAT de non-ruminant. L’utilisation de PAT de ruminant dans l’alimentation reste totalement interdite. Afin d’étendre la réintroduction des PATs de volaille dans les aliments pour porc et de PATs de porc dans les aliments pour volaille comme décidé récemment, des tests de détection visant les espèces de volaille et le porc étaient nécessaires pour vérifier qu’il n’y a pas de recyclage intra-spécifique.
Objectifs. Dans cette étude, nous décrivons la méthode de PCR en temps réel officielle de l’EURL-AP pour la détection simultanée du poulet (Gallus gallus L.) et de la dinde (Meleagris gallopavo L.), les deux espèces de volaille les plus utilisées.
Méthode. Les critères de qualité classiques pour la validation d’une méthode PCR sont considérés. Un seuil (valeur de cut-off) est requis pour l’interprétation des résultats. La transférabilité a été testée par une étude interlaboratoire.
Résultats. La méthode PCR développée amplifie un fragment de 84 pb situé sur les séquences de l'ARN ribosomal 12S, d'ARNt-Val et d'ARN ribosomal 16S de l’ADN mitochondrial. La méthode qualitative a été évaluée avec succès sur plusieurs critères de performance : spécificité, sensibilité, efficience et robustesse. L’applicabilité du test a été vérifiée sur des PATs de volaille et sur des aliments composés contenant 0,1 % en fraction massique de PAT de volaille. Une validation interlaboratoire a également démontré que la méthode de PCR en temps réel proposée est transférable.
Conclusions. La méthode PCR en temps réel combinant la détection du poulet et de la dinde est adaptée à la détection des PATs de volaille.
Abstract
Description of the subject. Use of all processed animal proteins (PAPs) in animal feeds was banned in the EU due to the outbreak of bovine spongiform encephalopathy (BSE). This total feed ban was progressively lifted for non-ruminant PAPs from 2013 onwards. Use of ruminant PAPs in feed remains totally prohibited. In order to extend the reintroduction of poultry PAPs in pig feed, and of pig PAPs in poultry feed, as recently decided, poultry species and pig detection tests were required to check that there is no intra-specific recycling.
Objectives. In this study, we describe the official EURL-AP real-time PCR method for the simultaneous detection of chicken (Gallus gallus L.) and turkey (Meleagris gallopavo L.), the two most widely used poultry species in PAPs.
Method. Classic quality criteria for the validation of a PCR method are considered. A cut-off is required for the interpretation of results. Transferability was tested through an interlaboratory study.
Results. The developed PCR assay amplifies an 84 bp fragment encompassing the mitochondrial DNA on 12S ribosomal RNA, tRNA-Val and 16S ribosomal RNA sequences. The qualitative method was successfully assessed for several performance criteria: specificity, sensitivity, efficiency and robustness. The applicability of the test was verified on poultry PAPs and on compound feed containing 0.1% in mass fraction of poultry PAPs. An interlaboratory validation study demonstrated that the proposed real-time PCR method is transferable.
Conclusions. The combined chicken-turkey real-time PCR assay is fit for the purpose of detecting poultry PAPs.
Table of content
Received 19 July 2023, accepted 29 January 2024, available online 19 February 2024.
This article is distributed under the terms and conditions of the CC-BY License (http://creativecommons.org/licenses/by/4.0)
1. Introduction
1Since the outbreak of bovine spongiform encephalopathy (BSE), the use of all processed animal proteins (PAPs) in animal feeds has been banned in the EU (European Commission, 2001; Olsvik et al., 2017).
2The validation of a real-time ruminant detection PCR test by the European Union Reference Laboratory for Animal Proteins in Feedingstuff (EURL-AP) to distinguish ruminant PAPs from other species has allowed this total feed ban, which was adopted in 2013 (Commission Regulation [EC] 56/2013), to be eased. It allowed recycling of non-ruminant PAPs from pig and poultry species to fish feed. Use of ruminant PAPs in feed remains totally prohibited (European Commission, 2013a).
3The European legislation gives different definitions to describe the term poultry depending on the usage (van Raamsdonk et al., 2019). In the context of processed animal proteins, the species included under the term poultry are chicken (Gallus gallus), turkey (Meleagris gallopavo), domestic ducks (mainly Anas platyrhynchos, Cairina moschata), domestic geese (mainly Anser sp.) and Helmeted guinea fowl (Numida meleagris) (European Commission, 2007; Scholtens et al., 2017; van Raamsdonk et al., 2019).
4Light microscopy and ruminant PCR analyses are applied for official control of feeds in the European Union, in order to verify the absence of proteins of ruminant origin (European Commission, 2013b; https://www.eurl.craw.eu). With the aim of extending this reintroduction of PAPs from poultry in pig feed, and of pig PAPs in poultry feed, as decided recently (European Commission, 2021), a test for detecting poultry species, as well as a pig detection test, are necessary to check that there is no intra-specific recycling.
5The detection and identification of species can be achieved using protein- or DNA-based methods (Fumière et al., 2009; Ghovvati et al., 2009), and several molecular approaches exist. The methods based on DNA analysis have many advantages. DNA is an extremely stable and long-lived biological molecule. Moreover, PCR-based methods are characterized by a high level of sensitivity, due to the exponential multiplication of the target. The use of a target present in multiple copies in cells also plays an important role for the sensitivity when the analyzed sample contains a low level of PAPs (Prado et al., 2009). For detection in processed products, the length of the amplicon must be short (Krcmar & Rencova, 2005; Fumière et al., 2006; Debode et al., 2007; Debode et al., 2017) because the DNA is fragmented due to mandatory processing conditions (European Commission, 2011).
6In order to align with the new regulation concerning the easing of the feed ban with poultry PAPs, a poultry PCR test has to be developed. This article describes the resulting practical PCR test for that purpose, based on the several constraints that needed to be taken into consideration.
2. Materials and methods
2.1. Real-time PCR method
7Primers and probe of the chicken-turkey PCR test developed were synthesized by Eurogentec (Seraing, Belgium). They have the following sequences: forward primer (Chic-Turk-F) 5’-TAGACTACCAAGGCGTAGCT-3’, reverse primer (Chic-Turk-R) 5’-AAGTCAAGGCGACCTTG-3’ and probe (Chic-Turk) 5’-AAAGCATTCAGCTTACACCTGAAA-3’. The probe was labelled with the reporter dye FAMTM (6-carboxyfluorescein) at the 5’ end and the quencher dye TAMRATM (Tetramethyl-6-Carboxyrhodamine) at the 3’ end.
8Real-time PCR was performed on a PCR platform combining a LightCycler 480 real-time PCR device (Roche Diagnostics, Mannheim, Germany) with 96-well reaction plates and the Universal Master Mix DMML-D2-D600 of Diagenode (Seraing, Belgium). A second PCR platform was used for the cut-off determination and the robustness evaluation. This second PCR platform combined a QuantStudio™ 6 Flex Real-time PCR system (Applied Biosystems, Foster city, CA, USA) with the Brilliant II QPCR Low ROX Master Mix (Agilent technologies, Santa Clara, CA, USA). The reaction mixture, with a total reaction volume of 25 µl, included Master Mix 1x, 0.9 µl of each primer (8.5 µM), 0.9 µl of probe (10 µM), double-distilled water to reach the volume of 20 µl, and 5 µl of DNA. Reaction mixtures were distributed on 96-well reaction plates that were developed for the specific thermocyclers. Once filled, a transparent adhesive film was affixed to the plate to close the wells and the plate was centrifuged (2 min at 500 rpm) prior to amplification in order to eliminate any air bubbles in the bottom of the wells. The thermal program included the following steps:
9– 50 °C for 2 min for the activation of UNG;
10– 95 °C for 10 min in order to inactivate the UNG, to activate the hot-start polymerase and to denature the DNA template;
11– 50 amplification cycles, with each cycle including a denaturation step at 95 °C for 15 s, followed by an annealing/elongation step at 50 °C for 60 s.
2.2. Specificity of the PCR method
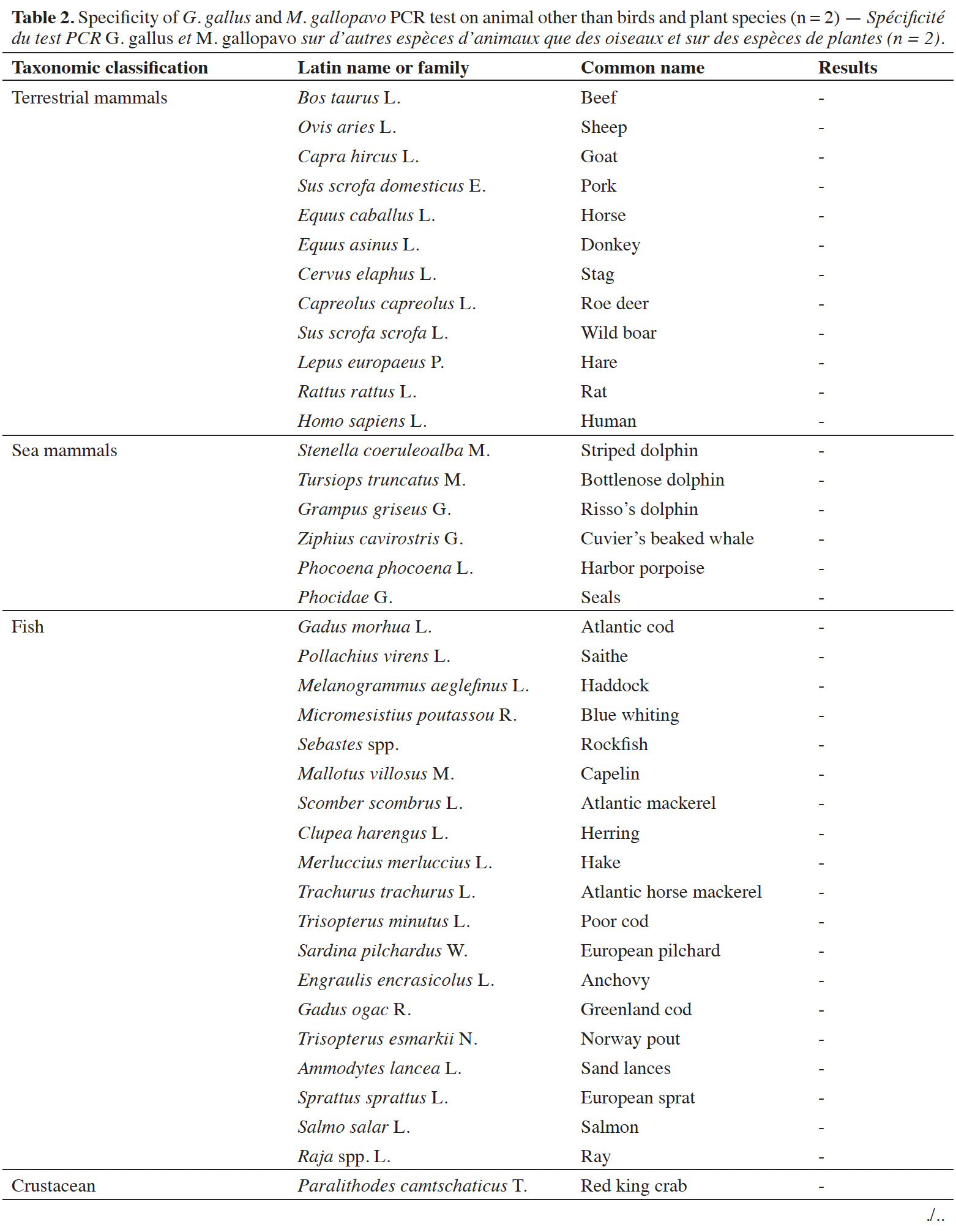
12The specificity of the chicken and turkey target was checked on a wide variety of animal and plant species. For the animal species, various categories were investigated, such as birds, fish, terrestrial and sea mammals. The possibility of a cross-reaction with human DNA was considered too. Ten ng of DNA were used in the reactions. Each DNA extract was tested at least in duplicate.
13Maximum precautions were taken to be sure of the purity of the tested DNA: most of the DNA were extracted from blood samples rather than meat, in order to avoid any contamination with DNA from other species. Where this was not possible, the DNA were extracted from meat. In this case, the outer area of the meat that may have been contaminated was eliminated and the test portions were collected in the central part of the samples. In the case of fish species, the skin was removed, and the extraction was carried out on muscle. The DNA extracts from pure species samples that were used for specificity testing were obtained via two extraction methods, depending on the type of matrix. The DNA from the blood samples were extracted with a “Qiagen Genomic DNA tip 20/G” kit (Qiagen GmbH, Hilden, Germany) from a test portion of 500 µl. Meanwhile, the DNA extracted from meat samples (test portion of 200 mg) were obtained via the CTAB method, in accordance with the protocol described in Annex A.3.1, which pertains to the ISO 21571:2005 international standard. The quantity and quality of the DNA extracts were evaluated with a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) by measuring the absorbance at 260 nm and 280 nm.
14The DNA extracts were diluted to obtain a concentration of 2 ng·µl-1. The amplifiability of the DNA extract (10 ng) was successfully checked via a real-time PCR, with the 18S target (Garikipati et al., 2006; Marien et al., 2018) for animals and the rbcL target (Debode et al., 2012) for plants.
2.3. PCR cut-off determination
15The cut-off value was determined on two PCR platforms, using calibration material provided by the Joint Research Centre (Geel, Belgium). This material consists of plasmid solutions with a plasmid bearing the PCR target issued from G. gallus, at three levels in copy number: 103, 24 and 8 copies·µl-1. To find out the cut-off value, the PCR method developed was performed on four PCR plates for each PCR platform, with four calibrations per plate, one calibration consisting of three replicates from three calibrant levels (nine wells) with 5 µl of calibrants in a reaction giving 515, 120 and 40 copies/well respectively (EURL-AP, 2022). The cut-off value is calculated using the Excel file available on the EURL-AP website for the poultry method (https://www.eurl.craw.eu/legal-sources-and-sops/method-of-reference-and-sops). The cut-off value in cycles is defined as the upper limit of the confidence interval of Cq values for 15 copies of the target (Olsvik et al., 2017). A quality criterion has been set for the cut-off value: it must exceed nine copies (EURL-AP, 2022).
2.4. Feed tested
16Several types of poultry feed matrices were used. The two blank poultry feeds used in the interlaboratory test were a starter feed for turkey (composition: maize, soybean oil cake, extruded full-fat soya, sunflower oil cake, limestone, monocalcium phosphate, lysine, vitamins and mineral premix, rapeseed oil, methionine, yeast, sodium chloride, sodium bicarbonate, choline chloride, veterinary drugs, threonine, feed enzymes) and a finishing feed for poultry (composition: wheat, barley, extruded soybean, dehulled sunflower seed oil cake, maize, wheat middlings, wheat bran, calcium carbonate, premix of additives, sodium chloride, sodium bicarbonate). The poultry feed with eggshells consisted of two broiler feeds (composition broiler feed 1: maize, dehulled soybean oil cake, wheat, animal fat, sorghum, dehulled sunflower seed oil cake, roasted soybeans, eggshells, inorganic dicalcium phosphate, sodium bicarbonate, sodium chloride and sodium butyrate; composition for broiler feed 2: maize, dehulled soybean oil cake, wheat, animal fat, sorghum, dehulled sunflower seed oil cake, eggshells, inorganic dicalcium phosphate and sodium chloride) and a turkey feed (composition: maize, wheat, dehulled soybean oil cake, animal fat, dehulled sunflower seed oil cake, inorganic dicalcium phosphate, dried eggshells, sodium bicarbonate and sodium chloride).
2.5. DNA extraction for industrial samples
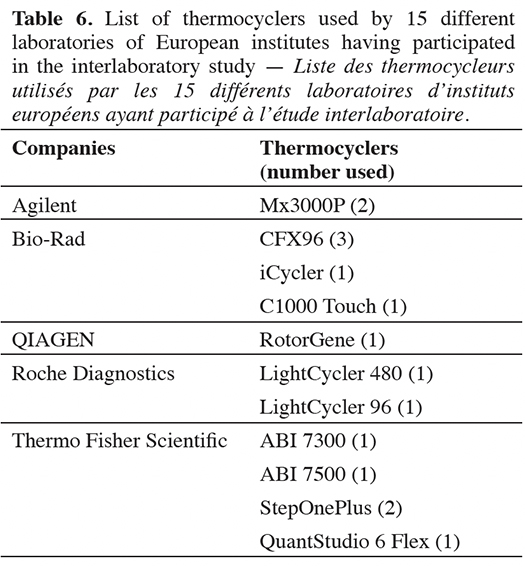
17The feed analysis for the PAP detection is regulated by European Commission Regulation (EU) No 51/2013 (European Commission, 2013b), and the mandatory DNA extraction method is described in the EURL-AP Standard Operating Procedure “DNA extraction using the Wizard® Magnetic DNA purification system for Food kit” (EURL-AP, 2014). This extraction method, which is based on the adaptation of the protocol for the Wizard Magnetic DNA Purification System for Food kit (Promega, Madison, USA), was used to extract DNA from feed, industrial PAPs and mixes at 0.1% in mass fraction of poultry PAPs in feed.
18In addition to the amplifiability tests described in the section “Specificity of the PCR method”, the industrial PAPs were tested for their purity with targets that were developed or evaluated within the framework of the EURL-AP activities (Marien et al., 2019; EURL-AP, 2021).
2.6. Cloning of the target and plasmid copy number determination and dilutions
19The 84 bp target from the G. gallus was cloned into the 3.9 kb pCR®2.1-TOPO plasmid vector (Invitrogen, Merelbeke, Belgium) according to the TOPO® TA Cloning® kit instructions (Invitrogen, Merelbeke, Belgium). The Genopure Plasmid Maxi Kit (Roche Diagnostics, Mannheim, Germany) was used to isolate plasmid DNA from bacterial cultures. The obtained plasmid DNA was linearized using the HindIII restriction enzyme (Roche Diagnostics GmbH, Mannheim, Germany), and then purified using phenol-chloroform-isoamyl alcohol extracts.
20The quantity and quality of plasmid DNA were measured using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies Inc., Wilmington, DE, USA) at 260 nm (A260) and 280 nm (A280) absorbance. The A260/A280 ratio makes it possible to determine the purity of DNA.
21The quantity of recovered plasmid DNA was converted into copy numbers as usual (Debode et al., 2010; Marien et al., 2018; Marien et al., 2022), with similar assumptions taken into consideration as previously (Marien et al., 2018):
22– 1 unit of absorbance at 260 nm corresponds to a concentration of 50 µg·ml-1;
23– the mean molar weight of one base pair is set at 635 Da.
24The sensitivity, efficiency and robustness of the PCR test were determined using diluted plasmid DNA. These dilutions were performed in water until an estimated copy number of 10,000 copies·5 µl-1 was reached. Higher dilutions of the target DNA were prepared in a solution containing 50 ng·µl-1 of salmon sperm DNA as background DNA. Low binding tubes were chosen to minimize DNA loss.
2.7. Limit of detection (LOD)
25Target sensitivity was evaluated as previously (Marien et al., 2018) following the recommendations of the former AFNOR XP V03-020-2 standard (AFNOR, 2003). This standard no longer exists, but the principles detailed in it remain valid. The absolute limit of detection (LODabs) was determined for the PCR assay (primers + probe + amplification program) on dilutions of plasmid material.
26The subsequent dilutions had to contain 50, 20, 10, 5, 2, 1 and 0.1 copies of the target respectively. Six PCRs had to be performed for each dilution. The LOD6 for the method is the smallest copy number for which the six PCRs were positive, but only if the highest dilution, which was supposed to contain the 0.1 copy, generated a maximum of one positive PCR signal on the six replicates per reaction. If more than one positive signal is observed for the 0.1 copy, then the DNA quantities have to be revised. The copy number corresponding to LOD6 is then tested 60 times on the same plate (determination of the LOD95%). The LOD95% is validated if at least 95% signals out of the 60 replicates are recorded as positive, corresponding to a minimum of 59 positive results. The highest acceptable copy number for LOD6 and LOD95% is 20 copies.
2.8. Efficiency
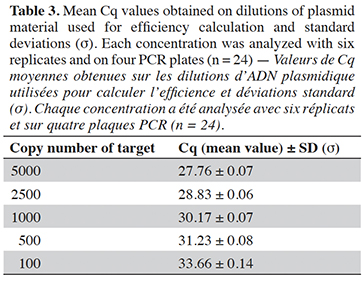
27The efficiency of the PCR assay was calculated as previously (Marien et al., 2022) using a series of dilutions of plasmid material at target levels of 5000, 2500, 1000, 500 and 100 copies. Each dilution was analyzed in six replicates and on four runs. The efficiency has to be between 90% to 110% (Broeders et al., 2014).
2.9. Robustness of the PCR method
28The robustness of the method was tested by slightly modifying the standard experimental conditions (CCMAS, 2010). Parameters considered were as usual (Broeders et al., 2014; Marien et al., 2018): the annealing temperature (50 °C +/- 1 °C), the primer or probe concentrations (standard or reduced by 30%) and the real-time PCR master mix volume (standard or +/- 1 µl), which involves a final reaction volume of 25 µl +/- 1 µl. Six replicates of the plasmid-borne target at 20 copies·5 µl-1 were tested under the various experimental conditions being considered. The robustness test was performed with real-time PCR platforms used for the cut-off determination. The acceptance criterion requires that any deviations from the standard protocol should not yield a negative result at the threshold of 20 copies of the target (Broeders et al., 2014).
2.10. Applicability of the PCR method
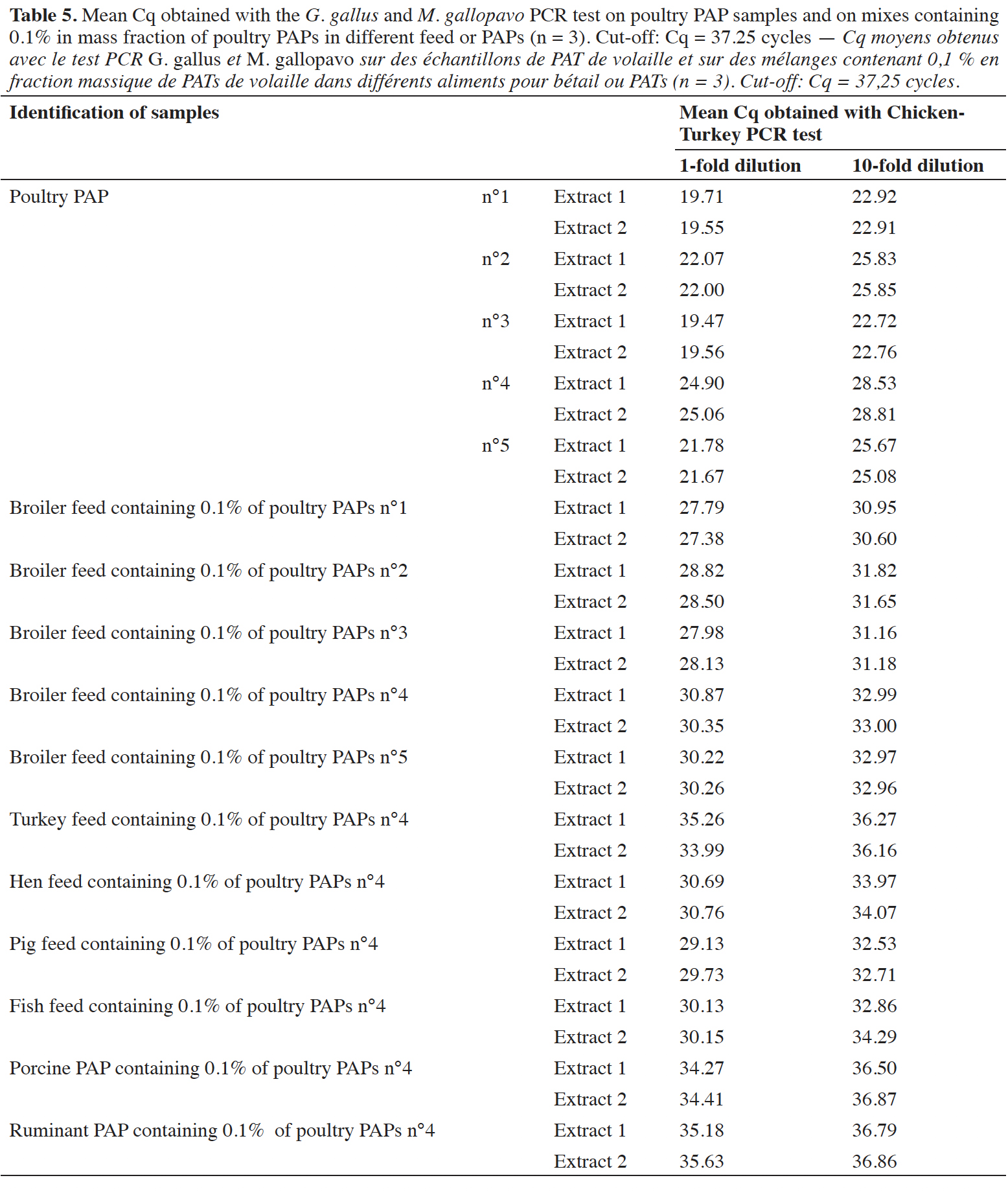
29The applicability of the PCR method was checked in triplicate on five poultry PAPs, which were submitted to various heat treatments in accordance with legislation, and on different feed mixes containing 0.1% in mass fraction of poultry PAPs. Different matrixes were used for these mixes: broiler feed, turkey feed, laying hen feed, pig feed, fish feed, pig PAPs and ruminant PAPs. These matrices were successfully verified as free of chicken and turkey DNA before being spiked with PAP meal. Two DNA extracts from each matrix, extracted with method described in the EURL-AP SOP (EURL-AP, 2014), were analyzed with the chicken-turkey PCR test and gave a negative result. Three feeds containing eggshells (authorized feed material) were also tested with the chicken-turkey PCR test. Four extracts of DNA were taken per feed sample, and each DNA extract was analyzed by PCR in triplicate.
2.11. Interlaboratory validation study
30Fifteen laboratories participated in the validation study. Each laboratory received ready-to-use reagents (primers and probe, the Universal Mastermix [Diagenode s.a.], the calibrants [provided by the JRC, Geel, Belgium]) for the cut-off determination, 10 blind DNA samples and the PCR negative controls made of PCR-grade water.
31The blind DNA samples were issued (Figure 1) from two poultry feeds, one consisting of a complete starter feed for fattening turkey and the other, a finishing feed for poultry. Poultry PAPs processed in accordance with method seven (temperature of at least 90 °C for 30 min on the cooking side, with drying treatment by heating at approximately 95 °C for 60 min – Annex, [European Commission, 2002]) was used to adulterate the first blank matrix at levels of 0.2, 0.1 and 0.04% in mass fraction. The targeted copy numbers in the reaction are +/- 500, 250 and 100 copies respectively. This was calculated with the help of the JRC calibrants.
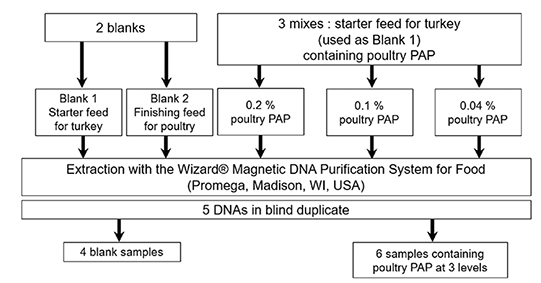
Figure 1. Production of the blind sample set used in the interlaboratory validation study of the real-time PCR method for the detection of poultry DNA in feedingstuffs. DNA were extracted according to the method described in the Standard Operating Procedure of EURL-AP (EURL-AP, 2014) — Production du set d’échantillons en aveugle utilisé dans l’étude de validation interlaboratoire de la méthode de PCR en temps réel pour la détection d’ADN de volaille dans les aliments pour bétail. Les ADN ont été extraits selon la méthode décrite dans la Procédure d’Opération Standard de l’EURL-AP (EURL-AP, 2014).
32These five samples were submitted to DNA extraction with the method recommended by EURL-AP at the EURL-AP, using the “Wizard® Magnetic DNA purification system for Food” kit (Promega, Madison, WI, USA) combined with the KingFisher Magnetic Particle Processor from ThermoFisher Scientific (Waltham, MA, USA) (EURL-AP, 2014).
33In order to provide all participants with the same DNA extracts, and to comply with the recommended extraction protocol, 65 extracts were prepared from 100 mg of each sample. All DNA extracts obtained for each level were analyzed. For the two blank samples, the DNA extracts were tested to verify the absence of PCR inhibition and contamination. For the adulterated samples, since the adulteration levels were low, the signals obtained from the various extracts showed a high degree of variability. That is why only extracts that were close to the targeted copy numbers were pooled for the 0.1 and 0.04% levels. For the 0.2% level, the number of copies obtained was higher than 500 copies for the majority of extracts. In order to obtain a level at +/- 500 copies, blank 1 DNA extract was added. The pooled DNA extracts were divided in vials of 250 µl.
34Finally, the sets of 10 blind DNA samples comprised five different kinds of samples in duplicate —two distinctive blank samples and three samples containing poultry PAPs at different concentrations in mass fraction: 0.2% in mass fraction equivalent to ~500 copies of the poultry target in the reaction, 0.1% in mass fraction (~250 copies of the poultry target in the reaction), 0.04% in mass fraction (~100 copies of the poultry target in the reaction). Each of the five kinds of samples was analyzed in 40 replicates, which were distributed on four different plates (10 replicates per plate). In order to avoid result biases due to an edge effect of the thermal block, samples from the same vial were placed in different places on each plate. On each of the four plates used to analyze samples, four calibrations consisting each of three replicates from three calibrant levels were also performed to fix the cut-off value (as described in “PCR cut-off determination”). The four PCR plates had to be performed within two or three consecutive days (the complete schemes for the plates are presented in Supplementary file 1).
35For each participant, a cut-off value in cycles, calculated at 15 copies, was automatically generated by the Excel file (https://www.eurl.craw.eu/legal-sources-and-sops/method-of-reference-and-sops/) provided by the organizer, once the file filled with the data of the calibrations. With the respective cut-off values, the replicates of the blind samples belonging to each participant were automatically ranked as positive or negative. A result for a well is considered as positive if the Cq value obtained for that well is smaller than the cut-off value determined for the platform of the laboratory in question.
36Before sending the material to the participants, the EURL-AP performed the full study with a randomly chosen set of samples, in order to verify that the obtained data satisfied expectations.
3. Results
37A small mitochondrial DNA sequence common to chicken and turkey was used as target for a real-time PCR test that enables the simultaneous specific detection of chicken and turkey. The chosen target has a small size (84 bp) and is located across 12S ribosomal RNA, tRNA-Val and 16S ribosomal RNA sequences. Some small sequence differences do exist between the two species, but the selected primers and probe show sequences that completely fit to both (Figure 2).

Figure 2. Alignment of the targeted gene portion amplified with the Chic-Turk-F and Chic-Turk-R primers on the reference mitochondrial sequences of G. gallus and M. gallopavo. The location of the primers and probe Chic-Turk is underligned — Alignement de la portion du gène ciblé amplifiée avec les amorces Chic-Turk-F et Chic-Turk-R sur les séquences mitochondriales de référence de G. gallus and M. gallopavo. La localisation des amorces et sonde Chic-Turk est soulignée.
38The specificity was tested on DNA from G. gallus and M. gallopavo, but also on nine other non-target bird species (Table 1). For non-target birds, poultry species besides chicken and turkey were considered, as well as Larus fuscus (black-backed gull), whose feces could be found in fish meals stored outside, as can be the case outside Europe. As expected, strong positive results were obtained only with G. gallus and M. gallopavo. However, late signals were observed with Phasianus colchicus, Columba livia and N. meleagris. These non-specific features should not cause problems because the amplification signals are late (Cq value > 35 cycles, therefore above a cut-off value), while 10 ng of DNA coming exclusively from these species are present in the reaction. Furthermore, the amplification curves obtained with C. livia show a non-optimal reaction efficiency (Figure 3).
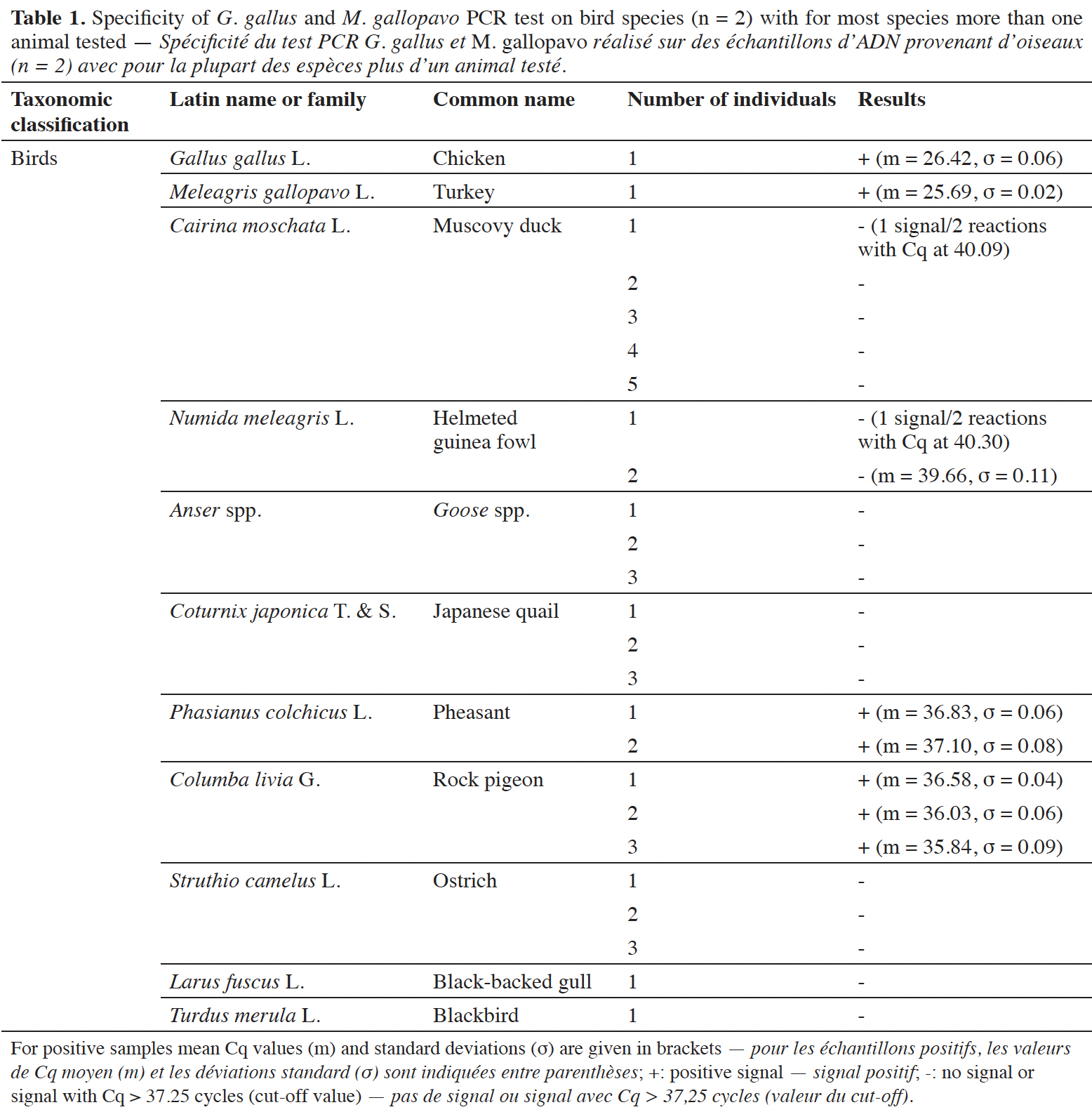
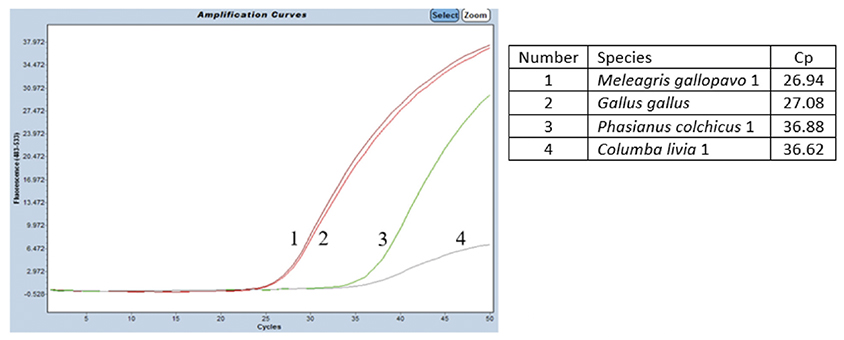 Figure 3. Amplification signals obtained on DNAs extracted from four different bird species using the chicken and turkey real-time PCR test. The PCR performed on the LightCycler 480 thermocycler (Roche Diagnostics Ltd) combined with the Universal Mastermix (Diagenode s.a.). Ten ng of DNA pure species per reaction. Cut-off: Cq = 37.25 cycles — Signaux d’amplification obtenus avec les extraits d’ADN de quatre espèces d’oiseaux différentes en utilisant le test PCR en temps réel poulet et dinde. PCR réalisée sur un thermocycleur LightCycler 480 (Roche Diagnostics Ltd) combiné avec l’Universal Mastermix (Diagenode s.a.). Dix ng d’ADN pure espèce par réaction. Cut-off: Cq = 37,25 cycles.
Figure 3. Amplification signals obtained on DNAs extracted from four different bird species using the chicken and turkey real-time PCR test. The PCR performed on the LightCycler 480 thermocycler (Roche Diagnostics Ltd) combined with the Universal Mastermix (Diagenode s.a.). Ten ng of DNA pure species per reaction. Cut-off: Cq = 37.25 cycles — Signaux d’amplification obtenus avec les extraits d’ADN de quatre espèces d’oiseaux différentes en utilisant le test PCR en temps réel poulet et dinde. PCR réalisée sur un thermocycleur LightCycler 480 (Roche Diagnostics Ltd) combiné avec l’Universal Mastermix (Diagenode s.a.). Dix ng d’ADN pure espèce par réaction. Cut-off: Cq = 37,25 cycles.
39A late and non-repeatable amplification curve was also obtained for C. moschata, but only with one individual out of five tested. No signal was obtained with the 38 other animal species (terrestrial mammals, sea mammals, fish and crustacean) and the seven plant species tested (Table 2).
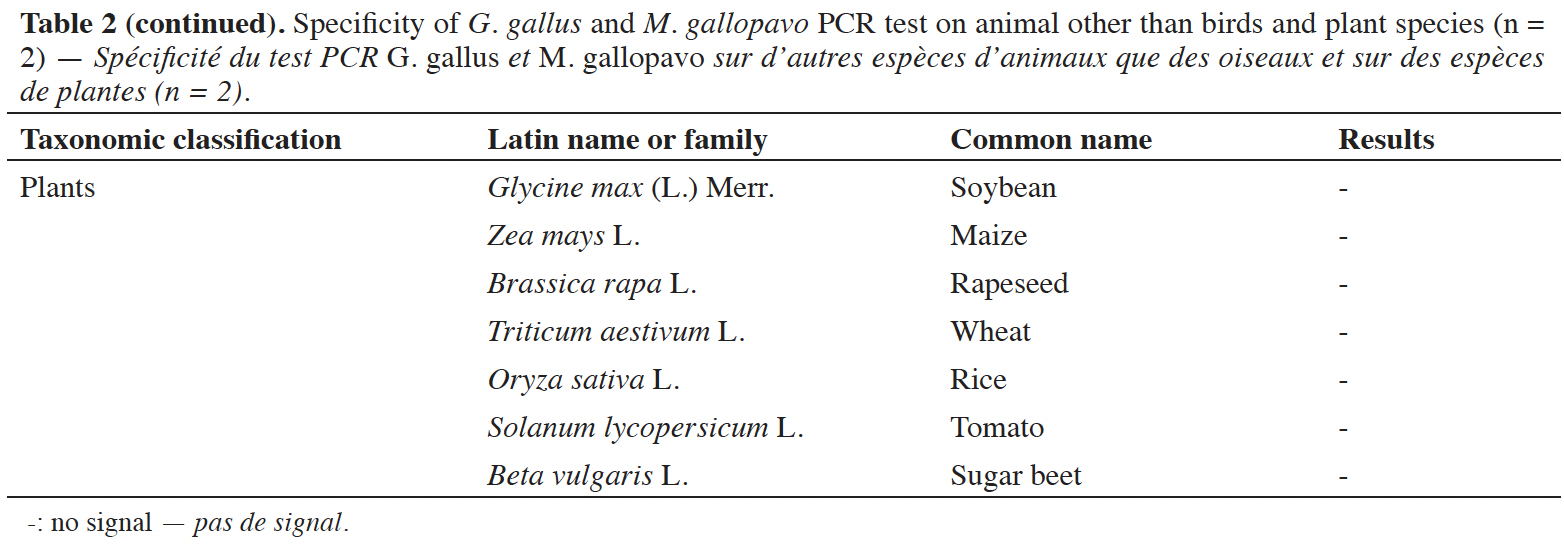 In order to interpret an amplification signal as a positive or negative result, a cut-off value is considered (EURL-AP, 2022). The Cq and copy numbers corresponding to the cut-off values were determined using two PCR platforms. The cut-off was chosen as the upper limit of the confidence interval of Cq values for 15 copies of the target. At this cut-off value, the late and non-repeatable signals obtained for N. meleagris and C. moschata are considered as negative results (Cq values after the cut-off value). The cut-off value obtained on the platform combining a LightCycler 480 (Roche Diagnostics) with the Universal Mastermix (Diagenode s.a.) is set at 37.25 cycles, which corresponds to 11.33 copies. For the second platform, a QuantStudio™ 6 (Applied Biosystems) combined with the Brilliant II QPCR Low ROX Master Mix (Agilent technologies), the cut-off value was calculated to 36.92 cycles, which corresponds to 11.27 copies. The cut-off values obtained for both platforms meet the quality criterion of more than nine copies.
In order to interpret an amplification signal as a positive or negative result, a cut-off value is considered (EURL-AP, 2022). The Cq and copy numbers corresponding to the cut-off values were determined using two PCR platforms. The cut-off was chosen as the upper limit of the confidence interval of Cq values for 15 copies of the target. At this cut-off value, the late and non-repeatable signals obtained for N. meleagris and C. moschata are considered as negative results (Cq values after the cut-off value). The cut-off value obtained on the platform combining a LightCycler 480 (Roche Diagnostics) with the Universal Mastermix (Diagenode s.a.) is set at 37.25 cycles, which corresponds to 11.33 copies. For the second platform, a QuantStudio™ 6 (Applied Biosystems) combined with the Brilliant II QPCR Low ROX Master Mix (Agilent technologies), the cut-off value was calculated to 36.92 cycles, which corresponds to 11.27 copies. The cut-off values obtained for both platforms meet the quality criterion of more than nine copies.
40Since the amplified fragment is a multicopy target, a plasmid containing the targeted DNA fragment was used, to have better control of the copy number, to perform sensitivity, efficiency and robustness tests. These three parameters reached the recommended acceptance criteria. Indeed, for sensitivity, the LOD6 was estimated at five copies following the former AFNOR XP V03-020-2 standard approach (AFNOR, 2003), and for the LOD95%, which was also tested at five copies, 60/60 positive signals were obtained.
41However, with a cut-off value set at 15 copies, the LOD95% was at 20 copies with 60/60 positive signals obtained. Therefore, the PCR test reaches the recommended performance criteria (≤ 20 copies).
42The PCR efficiency in a range from 100 to 5,000 copies was evaluated at 94.2% (Table 3). The efficiency calculated per plate was also always higher than 90% and therefore met the acceptance criterion set by Broeders et al. (2014).
43The robustness of the PCR method has been demonstrated, even when the cut-off value is calculated at 15 copies. Positive results have been obtained in all tested deviations (Table 4) to the standard protocol at a target copy number of 20 for the PCR.
44Positive signals were also obtained on five different poultry PAPs (pure meals), demonstrating the applicability of the PCR test on real-life samples (Table 5). These poultry PAPs were used to prepare a mix at 0.1% in mass fraction of poultry PAPs in a broiler feed. Six mixes were also prepared in different matrices at the level of 0.1% m·m-1, with poultry PAPs number 4 giving the latest signals (Cq ~25 cycles). All mixes at 0.1% were detected as containing chicken and/or turkey DNA (Table 5). A matrix effect was observed, with a difference of six cycles between the Cq values obtained for a single kind of PAPs in different matrices. This effect does not impact detection. The tenfold dilutions demonstrated that there is no inhibition with pure poultry PAPs. For the mixes at 0.1%, a slight inhibitory effect was observed on the amplification of the G. gallus and M. gallopavo target, depending on the matrices.
45Three feeds containing eggshells were also tested. Eggshells are authorized for feed. Although DNA extraction from eggs gives a low yield of collected DNA (Rikimaru & Takahashi, 2009), it was important to prove that the presence of this authorized feed material in poultry feed does not interfere with the results. Of the three feeds tested, two were negative and one showed only traces of chicken-turkey DNA. As can be seen in figure 4, the obtained amplification curves show late signals with a very low efficiency.
 Figure 4. Amplification signals obtained on DNAs extracted from a sample containing eggshells (only one feed sample out of the three containing eggshells gives rise to late signals) with the chicken and turkey real-time PCR test. The PCR performed on the LightCycler 480 thermocycler (Roche Diagnostics Ltd) combined with the Universal Mastermix (Diagenode s.a.). Five µl of DNA extract were analyzed per reaction and three replicates by DNA extract. Cut-off: Cq = 37.25 cycles — Signaux d’amplification obtenus avec des extraits d’ADN d’un échantillon contenant des coquilles d’œufs (seulement un échantillon d’aliment sur les trois contenant des coquilles d’œufs donnent des signaux tardifs) avec le test PCR en temps réel poulet et dinde. PCR réalisée sur un thermocycleur LightCycler 480 (Roche Diagnostics Ltd) combiné avec l’Universal Mastermix (Diagenode s.a.). Cinq µl d’extrait d’ADN ont été analysés par réaction et trois réplicats par extrait d’ADN. Cut-off: Cq = 37,25 cycles.
Figure 4. Amplification signals obtained on DNAs extracted from a sample containing eggshells (only one feed sample out of the three containing eggshells gives rise to late signals) with the chicken and turkey real-time PCR test. The PCR performed on the LightCycler 480 thermocycler (Roche Diagnostics Ltd) combined with the Universal Mastermix (Diagenode s.a.). Five µl of DNA extract were analyzed per reaction and three replicates by DNA extract. Cut-off: Cq = 37.25 cycles — Signaux d’amplification obtenus avec des extraits d’ADN d’un échantillon contenant des coquilles d’œufs (seulement un échantillon d’aliment sur les trois contenant des coquilles d’œufs donnent des signaux tardifs) avec le test PCR en temps réel poulet et dinde. PCR réalisée sur un thermocycleur LightCycler 480 (Roche Diagnostics Ltd) combiné avec l’Universal Mastermix (Diagenode s.a.). Cinq µl d’extrait d’ADN ont été analysés par réaction et trois réplicats par extrait d’ADN. Cut-off: Cq = 37,25 cycles.
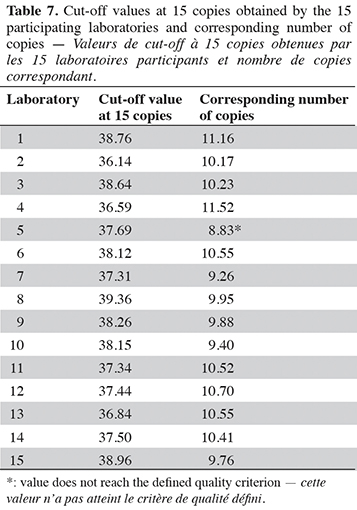
46Fifteen different laboratories within European institutes participated in the interlaboratory validation study, with 11 types of thermocyclers from five major companies (Table 6). Out of the 15 cut-off values calculated, only one does not meet the quality criterion fixed by the organizer. The 14 valid cut-off values range from 36.14 to 39.36 cycles (Table 7).
47Blind samples were tested 40 times per laboratory (10 replicates x 2 samples x 2 runs). The rates of false-positive and false-negative results were calculated per laboratory and for all the results (Table 8). Across all results, the global rate of false-positive results is 1.83% (22/1200) and the global rate of false-negative results is 0.22% (33/1800). It must be emphasized that all the false-positive results were recorded by the same lab (lab 3). At the level of 0.2% of poultry PAPs in feeding stuffs, only one reaction out of 600 gave a negative result. For the level at 0.1% of poultry PAPs in feeding stuffs, no false-negative result was recorded out of 600 reactions. Given that the 0.1% level provides a detection rate of 100%, and that this level is lower than the 0.2%, the negative result for the latter is probably due to an oversight or pipetting error. For the level at 0.04% of poultry PAPs in feeding stuffs, three reactions out of 600 gave a negative result; this corresponds to an error rate of 0.5% for this level.
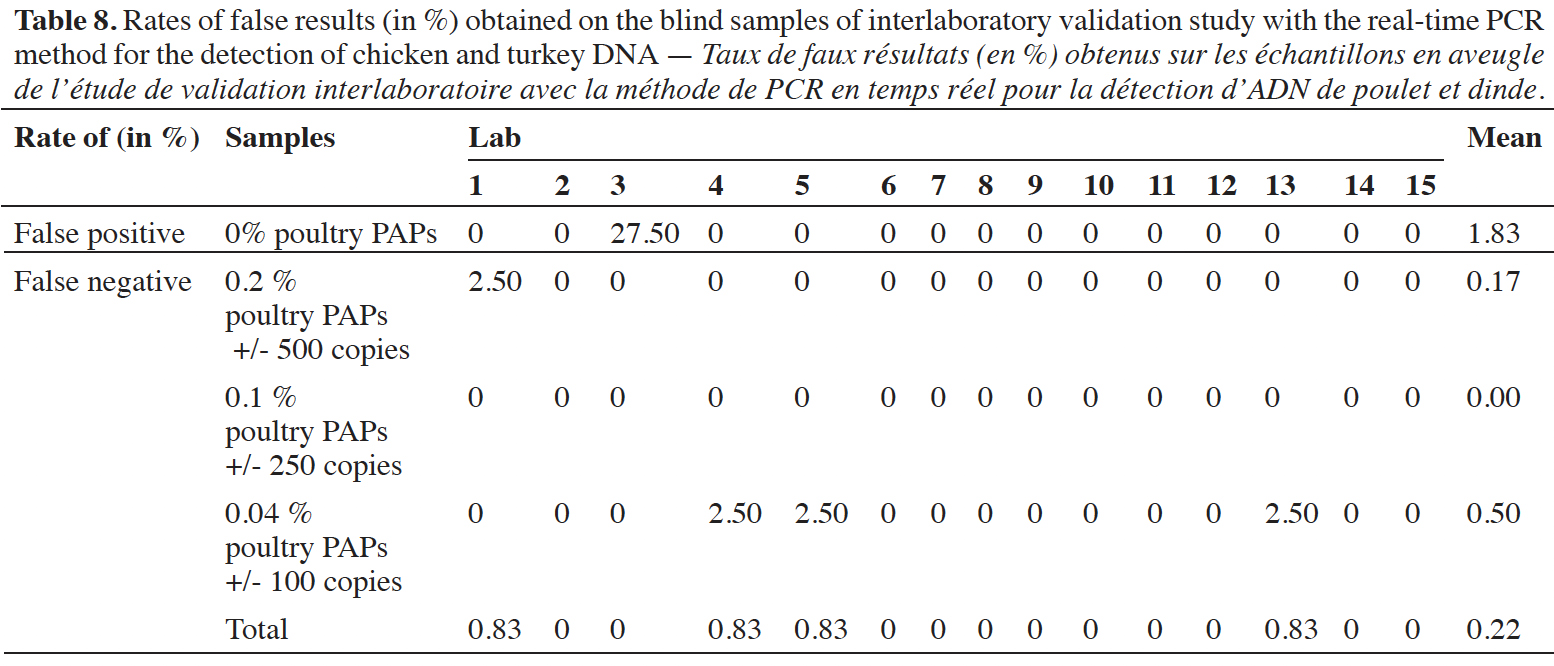
48These values are far below an error rate of 5%, and the method can therefore be considered as fit for purpose. The interlaboratory validation study has demonstrated that the protocol for chicken and turkey detection is transferable.
4. Discussion
49The reintroduction of PAPs from poultry in pig feed (European Commission, 2021) implies having a test allowing the detection of poultry species in order to check that there is no intra-specific recycling, no presence of poultry PAPs in poultry feed. However, the design of a PCR test for the detection of only poultry species, as defined in Regulation (EC) No 1234/2007, is difficult given the taxonomic diversity of the species that are legally included under this umbrella. It includes chicken (G. gallus), turkey (M. gallopavo), domestic ducks (mainly A. platyrhynchos, C. moschata), domestic geese (mainly Anser sp.) and Helmeted guinea fowl (N. meleagris) (European Commission, 2007; Scholtens et al., 2017; van Raamsdonk et al., 2019). Most of the PCR tests developed prior to this point usually detect only one poultry species, as in species-specific PCR assays (Krcmar & Rencova, 2005; Laube et al., 2007; Fumière et al., 2010; Pegels et al., 2012; Amaral et al., 2015; Ren et al., 2017; Xiang et al., 2017; Kim & Kim, 2018; Wang et al., 2021). Nevertheless, some tests were combined by performing them in multiplex (Köppel et al., 2008; Ng et al., 2012; Cheng et al., 2014; Hou et al., 2015; Scholtens et al., 2017; Kim & Kim, 2019; Salam et al., 2022). It should, however, be stressed that, for some of these assays, the specificity assessment is extremely limited, particularly at the level of the species of birds being tested. Other tests identify any avian species, such as the real-time PCR assay published by Pegels et al. (2014) or the PCR-RFLP technique described by Stamoulis et al. (2010), but these methods are not specific enough for the purpose of PAP detection. It is the method published by Scholtens et al. (2017) that is best suited to this purpose. However, as it is made from a combination of two targets, one for chicken and turkey and another one for geese and ducks, it is difficult to use it in a test with a calibration curve that has to be devoted to a single target.
50From discussions with EFPRA (European Fat Processors and Renderers Association), it emerged that chicken (G. gallus) and turkey (M. gallopavo) represent, together with mulard ducks (C. moschata x A. platyrhynchos), the main sources of poultry PAPs, which will therefore almost always contain either chicken or turkey material in combination with other poultry species. Nevertheless, some rendering plants may produce pure mulard duck PAPs. This is a high-value feed material, which is valued at a better price in pet food. Therefore, it will almost never be used as a single poultry species PAPs in feed. Thus, in practice, having a well-designed chicken-turkey PCR test is sufficient to allow efficient detection of poultry PAPs in feed.
51This study proposes a real-time PCR method, based on a small mitochondrial DNA target, that enables the simultaneous detection of chicken and turkey specifically.
52The small size of the target and its location on the mitochondrial DNA have the advantage of allowing detection of poultry DNA from chicken and turkey at low levels, even in a highly processed product. Indeed, a mitochondrion contains several copies of its genome, and several mitochondria can be present in a single cell (Marien et al., 2018; Cavelier et al., 2000). However, this multicopy trait is a disadvantage for quantitative purposes, as the copy number per cell varies depending on the tissue in question (Marien et al., 2018).
53The specificity tested experimentally gives good results with respect to the animal and plant species tested. Only two common bird species give an aspecific reaction. These non-specific features encountered with two common bird species should not cause problems because even though the high amount of DNA used, the amplification signals obtained are late and close to the cut-off value.
54In silico analysis, however, showed that the PCR test would provide positive results with other representatives of the genera Gallus and Meleagris (Supplementary file 2). Extensive comparisons, especially with other Galliformes (Supplementary file 3), Anseriformes (Supplementary file 4), Columbiformes (Supplementary file 5), Passeriformes (Supplementary file 6), Gruiformes (Supplementary file 7) and Charadriiformes (Supplementary file 8), showed a limited risk of interference except for three Galliform species: Francolinus pintadeanus, Lagopus muta japonica and Tragopan temminckii. However, these three species are far less common than chicken and turkey. The specificity is therefore fit for the purpose.
55The other performance criteria, which are the sensitivity, the efficiency and the robustness were also reached. The applicability of the PCR test has been demonstrated during this study on poultry PAPs and on compound feed containing 0.1% in mass fraction of poultry PAPs. The applicability of the PCR test has also been demonstrated in the publication by Axmann et al. (2015).
56Finally, an interlaboratory validation study showed that the method proposed is transferable.
5. Conclusions
57In brief, the design of a real poultry PCR assay is difficult, as the term ‘poultry’ acts as an umbrella for taxonomically unrelated species. This is why preference was given to a chicken-turkey PCR test. Indeed, chicken (G. gallus) and turkey (M. gallopavo) represent the main sources of poultry PAPs.
58The developed PCR assay targets an 84 bp fragment located in mitochondrial DNA across the 12S ribosomal RNA, tRNA-Val and 16S ribosomal RNA sequences. The specificity produces good results with respect to the animal and plant species tested. Only two common bird species showed a lack of specificity but with late signals close to the cut-off value. Taking into account the aim for which the test is designed, the risk of interference is extremely reduced. Due to the multicopy nature of the target, the assay is very sensitive for poultry PAP detection. Besides sensitivity (LOD6 and LOD95%), efficiency and robustness also met the required acceptance criteria. Furthermore, it was checked that the method is applicable to real-life samples from industry, even with a level of 0.1% in mass fraction of poultry PAPs in feed, while the presence of eggshells as material in the feed does not really interfere with the results. Moreover, the interlaboratory validation study has demonstrated that the method is transferable. Therefore, the conclusion is that the PCR assay developed herein is fit for the purpose of poultry PAP detection.
Acknowledgements
59The authors would like to thank the laboratories that participated in the interlaboratory validation study: Agroscope, Posieux, Switzerland; Animal and Plant Health Agency (APHA-Penrith), Penrith, United Kingdom; Cyprus Veterinary Services, Nicosia, Cyprus; Danish Veterinary and Food Administration, Ringsted, Denmark; Darling Ingredients Nederland BV, Son, The Netherlands; Federal Institute for Risk Assessment (BfR), Berlin, Germany; Instituto Nacional de Investigação Agrária e Veterinária (INIAV), Oeiras, Portugal; Istituto Zooprofilattico Sperimentale del Piemonte, Liguria e Valle d'Aosta (IZSTO – CreAA), Torino, Italy; National Diagnostic Centre of Food and Veterinary Service, Ljubljana, Slovenia; National Diagnostic Research Veterinary Medical Institute, Sofia, Bulgaria; National Institute of Nutrition and Seafood Research (NIFES), Bergen, Norway; NutriControl BV, Veghel, The Netherlands; Österreichische Agentur für Gesundheit und Ernährungssicherheit (AGES GmbH), Linz, Austria; RIKILT Wageningen University & Research, Wageningen, The Netherlands; Service Commun des Laboratoires du MINEFI, Rennes, France. The authors are grateful to Philippe Corbisier, Janka Mátrai and Stéphane Mazoua (EC JRC IRMM, Geel, Belgium) for the production of the calibrants and Christoph von Holst (JRC Geel, Belgium) for statistics and fruitful discussions. The authors would like to thank also Julie Hulin and Cécile Ancion (of the Molecular biology team, Unit 12, CRA-W) for efficient technical assistance.
Bibliographie
AFNOR, 2003. Détection et quantification des organismes végétaux génétiquement modifiés et produits dérivés. Partie 2 : méthodes basées sur la réaction de polymérisation en chaîne. Saint-Denis La Plaine, France : AFNOR; Report No.: AFNOR Standard XP-V-03-020-2.
Amaral J.S. et al., 2015. Identification of duck, partridge, pheasant, quail, chicken and turkey meats by species-specific PCR assays to assess the authenticity of traditional game meat Alheira sausages. Food Control, 47, 190-195, doi.org/10.1016/j.foodcont.2014.07.009
Axmann S. et al., 2015. Species identification of processed animal proteins (PAPs) in animal feed containing feed materials from animal origin. Food Addit. Contam., Part A, 32(7), 1089-1098, doi.org/10.1080/19440049.2015.1036321
Broeders S. et al., 2014. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol., 37, 115-126, doi.org/10.1016/j.tifs.2014.03.008
Cavelier L., Johannisson A. & Gyllensten U., 2000. Analysis of mtDNA copy number and composition of single mitochondrial particles using flow cytometry and PCR. Exp. Cell Res., 259, 79-85, doi.org/10.1006/excr.2000.4949
CCMAS (Codex Committee on Methods of Analysis and Sampling), 2010. Guidelines on performance criteria and validation of methods for detection, identification and quantification for specific DNA sequences and specific proteins in foods (Rep. No. CAC/GL 74-2010). Roma : FAO-WHO.
Cheng X. et al., 2014. Multiplex real-time PCR for the identification and quantification of DNA from duck, pig and chicken in Chinese blood curds. Food Res. Int., 60, 30-37, doi.org/10.1016/j.foodres.2014.01.047
Debode F., Janssen E. & Berben G., 2007. Physical degradation of genomic DNA of soybean flours does not impair relative quantification of its transgenic content. Eur. Food Res. Technol., 226, 273-280, doi.org/10.1007/s00217-006-0536-1
Debode F., Marien A., Janssen E. & Berben G., 2010. Design of multiplex calibrant plasmids, their use in GMO detection and the limit of their applicability for quantitative purposes owing to competition effects. Anal. Bioanal. Chem., 396, 2151-2164, doi.org/10.1007/s00216-009-3396-2
Debode F., Janssen E., Marien A. & Berben G., 2012. DNA detection by conventional and real-time PCR after extraction from vegetable oils. J. Am. Oil Chem. Soc., 89, 1249-1257, doi.org/10.1007/s11746-012-2007-0
Debode F. et al., 2017. The influence of amplicon length on real-time PCR results. Biotechnol. Agron. Soc. Environ., 21, 3-11, doi.org/10.25518/1780-4507.13461
EURL-AP, 2014. Standard Operating Procedure, version 1.1. DNA extraction using the “Wizard Magnetic DNA purification system for food” kit. Gembloux, Belgium: CRA-W, https://www.eurl.craw.eu/legal-sources-and-sops/method-of-reference-and-sops/, (01/08/2014).
EURL-AP, 2021. Standard Operating Procedure. Detection of ruminant DNA in feed using real-time PCR. Gembloux, Belgium: CRA-W, https://www.eurl.craw.eu/legal-sources-and-sops/method-of-reference-and-sops/, (01/10/2021).
EURL-AP, 2022. Poultry-Chicken-Turkey-Cut-off-at-15-copies-V1.0(1).xlsx. Gembloux, Belgium: CRA-W, https://www.eurl.craw.eu/legal-sources-and-sops/method-of-reference-and-sops, (01/08/2022).
EURL-AP, 2022. Standard Operating Procedure. Detection of poultry (chicken and turkey) DNA in feed using real-time PCR. Gembloux, Belgium: CRA-W, https://www.eurl.craw.eu/legal-sources-and-sops/method-of-reference-and-sops/, (01/08/2022).
European Commission, 2001. Regulation (EC) No 999/2001 of the European Parliament and the council of 22 May 2001 laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies. Off. J. Eur. Union, L147, 31/05/2001, 1-40.
European Commission, 2002. Regulation (EC) No 1774/2002 of the European Parliament and of the Council of 3 October 2002 laying down health rules concerning animal by-products not intended for human consumption. Off. J. Eur. Union, L273/1, 10/10/2002, 1.
European Commission, 2007. Council regulation (EC) No 1234/2007 of 22 October 2007 establishing a common organisation of agricultural markets and on specific provisions for certain agricultural products (Single CMO Regulation). Off. J. Eur. Union, L299, 16/11/2007, 1-149.
European Commission, 2011. Commission regulation (EU) No 142/2011. Implementing regulation (EC) No 1069/2009 of the European Parliament and of the Council laying down health rules as regards animal by-products and derived products not intended for human consumption and implementing Council Directive 97/78/EC as regards certain samples and items exempt from veterinary checks at the border under that Directive. Off. J. Eur. Union, L54, 26/02/2011, 1-254.
European Commission, 2013a. Commission regulation (EU) 2013/56. Amending Annexes I and IV to Regulation (EC) No 999/2001 of the European Parliament and of the Council laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies. Off. J. Eur. Union, L21, 24/01/2013, 3-16.
European Commission, 2013b. Commission regulation (EU) 2013/51. Amending regulation (EC) No 152/2009 as regards the methods of analysis for the determination of constituents of animal origin for the official control of feed. Off. J. Eur. Union, L20, 23/01/2013, 33-43.
European Commission, 2021. Commission regulation (EU) 2021/1372 of 17 August 2021 amending Annex IV to Regulation (EC) No 999/2001 of the European Parliament and of the Council as regards the prohibition to feed non-ruminant farmed animals, other than fur animals, with protein derived from animals. Off. J. Eur. Union, L295, 18/08/2021, 1-17.
Fumière O. et al., 2006. Effective PCR detection of animal species in highly processed animal byproducts and compound feeds. Anal. Bioanal. Chem., 385, 1045-1054, doi.org/10.1007/s00216-006-0533-z
Fumière O. et al., 2009. Methods of detection, species identification and quantification of processed animal proteins in feedingstuffs. Biotechnol. Agron. Soc. Environ., 13, 59-70.
Fumière O. et al., 2010. Development of a real-time PCR protocol for the species origin confirmation of isolated animal particles detected by NIRM. Food Addit. Contam., Part A, 27(8), 1118-1127, doi.org/10.1080/19440049.2010.481639
Garikipati D.K., Gahr S.A. & Rodgers B.D., 2006. Identification, characterization, and quantitative expression analysis of rainbow trout myostatin-1a and myostatin-1b genes. J. Endocrinol., 190, 879-888, doi.org/10.1677/joe.1.06866
Ghovvati S. et al., 2009. Fraud identification in industrial meat products by multiplex PCR assay. Food Control, 20, 696-699, doi.org/10.1016/j.foodcont.2008.09.002
Hou B. et al., 2015. Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Sci., 101, 90-94, doi.org/10.1016/j.meatsci.2014.11.007.
ISO 21571:2005. Foodstuffs – Methods of analysis for the detection of genetically modified organisms and derived products – Nucleic acid extraction. Geneva, Switzerland: International Organisation for Standardization.
Kim M.-J. & Kim H.-Y., 2018. Development of a fast duplex real-time PCR assay for simultaneous detection of chicken and pigeon in raw and heat-treated meats. Food Control, 85, 1-5, doi.org/10.1016/j.foodcont.2017.09.012
Kim M.-J. & Kim H.-Y., 2019. A fast multiplex real-time PCR assay for simultaneous detection of pork, chicken, and beef in commercial processed meat products. Food Sci. Technol., 114, 108390, doi.org/10.1016/j.lwt.2019.108390
Köppel R., Ruf J., Zimmerli F. & Breitenmoser A., 2008. Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, chicken and turkey. Eur. Food Res. Technol., 227, 1199-1203, doi.org/10.1007/s00217-008-0837-7
Krcmar P. & Rencova E., 2005. Quantitative detection of species-specific DNA in feedstuffs and fish meals. J. Food Prot., 68(6), 1217-1221.
Laube I. et al., 2007. Development and design of a ‘ready-to-use’ reaction plate for a PCR-based simultaneous detection of animal species used in foods. Int. J. Food Sci. Technol., 42, 9-17, doi.org/10.1111/j.1365-2621.2006.01154.x
Marien A. et al., 2018. Detection of Hermetia illucens by real-time PCR. J. Insects Food Feed, 4(2), 115-122, doi.org/10.3920/JIFF2017.0069
Marien A. et al., 2019. The horse meat scandal – The European analytical response. In: Burns M., Foster L. & Walker M., eds. DNA techniques to verify food authenticity: applications in food fraud. Chapter 16. Cambridge, UK: Royal Society of Chemistry, 177-188, doi: 10.1039/9781788016025
Marien A. et al., 2022. Detection of Alphitobius diaperinus by real-time polymerase chain reaction with a single-copy gene target. Front. Vet. Sci., 9, 718806, doi.org/10.3389/fvets.2022.718806
Ng J., Satkoski J., Premasuthan A. & Kanthaswamy S., 2012. A nuclear DNA-based species determination and DNA quantification assay for common poultry species. J. Food Sci. Technol., 51(12), 4060-4065, doi.org/10.1007/s13197-012-0893-7
Olsvik P.A. et al., 2017. Multi-laboratory evaluation of a PCR method for detection of ruminant DNA in commercial processed animal proteins. Food Control, 73, Part B, 140-146, doi.org/10.1016/j.foodcont.2016.07.041
Pegels N. et al., 2012. Evaluation of a TaqMan real-time PCR assay for detection of chicken, turkey, duck, and goose material in highly processed industrial feed samples. Poult. Sci., 91, 1709-1719, doi.org/10.3382/ps.2011-01954
Pegels N., González I., García T. & Martín R., 2014. Avian-specific real-time PCR assays for authenticity control in farm animal feeds and pet foods. Food Chem., 142, 39-47, doi.org/10.1016/j.foodchem.2013.07.031
Prado M. et al., 2009. Novel approach for interlaboratory transfer of real-time PCR methods: detecting bovine meat and bone meal in feed. Anal. Bioanal. Chem., 394, 1423-1431, doi.org/10.1007/s00216-009-2796-7
Ren Y. et al., 2017. A novel quantitative real-time PCR method for identification and quantification of mammalian and poultry species in foods. Food Control, 76, 42-51, doi.org/10.1016/j.foodcont.2017.01.003
Rikimaru K. & Takahashi H., 2009. A simple and efficient method for extraction of PCR amplifiable DNA from chicken eggshells. Anim. Sci. J., 80(2), 220-223, doi.org/10.1111/j.1740-0929.2008.00624.x
Salam M.R. et al., 2022. Detection of chicken and turkey in different beef matrix by species-specific multiplex PCR assay. Sci. Afr., 17, e01338, doi.org/10.1016/j.sciaf.2022.e01338
Scholtens I.M.J., Prins T.W. & van Raamsdonk L.W.D., 2017. Specificity of a novel TaqMan PCR method for detection of poultry DNA. Food Control, 73, Part B, 532-539, doi.org/10.1016/j.foodcont.2016.08.045
Stamoulis P., Stamatis C., Sarafidou T. & Mamuris Z., 2010. Development and application of molecular markers for poultry meat identification in food chain. Food Control, 21, 1061-1065, doi.org/10.1016/j.foodcont.2009.12.027
van Raamsdonk L.W.D. et al., 2019. Bridging legal requirements and analytical methods: a review of monitoring opportunities of animal proteins in feed. Food Addit. Contam., Part A, 36, 46-73, doi.org/10.1080/19440049.2018.1543956
Wang W. et al., 2021. A novel quantitative real-time PCR method for the detection of mammalian and poultry species based on a shared single-copy nuclear DNA sequence. Food Chem., 341(2), 128170, doi.org/10.1016/j.foodchem.2020.128170
Xiang W. et al., 2017. Identification of a chicken (Gallus gallus) endogenous reference gene (Actb) and its application in meat adulteration. Food Chem., 234, 472-478, doi.org/10.1016/j.foodchem.2017.05.038






