- Accueil
- volume 13 (2009)
- numéro spécial
- Methods of detection, species identification and quantification of processed animal proteins in feedingstuffs
Visualisation(s): 0 (0 ULiège)
Téléchargement(s): 0 (0 ULiège)
Methods of detection, species identification and quantification of processed animal proteins in feedingstuffs

Abstract
The ban of processed animal proteins (PAPs) in feed for farmed animals led to a significant reduction of the number of bovine spongiform encephalopathy cases. Presently, optical microscopy remains the only reference method for the detection of PAPs to be applied for official control as required by Commission Directive 2003/126/EC. The legislation also foresees that other methods may be applied in addition to classical microscopy, if – for instance – they provide more information about the origin of the animal constituents. Therefore, alternative and complementary techniques were developed as such or in combination. The most promising ones seem to be PCR (Polymerase Chain Reaction), near infrared microscopy and imaging, as well as immunology. Within the framework of a PAP ban regardless of its species origin (total feed ban), most of the studies were mainly focused on the ability of the techniques to detect the presence of PAPs at 0.1% (mass percentage of constituents of animal origin in feed) as indicated as limit of detection in the official method protocol. A possible modification of the legislation requires that the techniques are also able to determine their species origin and to quantify them. The present paper gives a state of the art of the different methods.
Table des matières
1. Introduction
1The outbreak of bovine spongiform encephalopathy (BSE) urged the European Union to take several decisions in order to avoid the transmission of its most probable causal agent through the food chain. At present, with exceptions for fish meal, processed animal proteins (PAPs) including meat and bone meals (MBM) are banned from use as feed ingredients for all farmed animals. Moreover, the use of PAPs is controlled within the European Union through several regulations. Regulation (EC) n°999/2001 prohibits explicitly the feeding of mammalian proteins to ruminants, whereas Regulation (EC) n°1774/2002 introduced several provisions, which are mainly:
2– the ban of feeding animals with proteins from the same species (ban of intra-species recycling),
3– the classification of animal by-products (ABPs) into 3 categories reflecting different safety levels and including the risk due to transmissible spongiform encephalopathy (TSE).
4Only material from category 3, i.e. that originates from animals fit for human consumption could be used to feed farm animals. Enforcing these regulations required analytical methods capable to allow species-specific identification. The lack of such methods led to the introduction of an extended feed ban for all farmed animals by amending Regulation (EC) n°999/2001 through Commission Regulation (EC) n°1234/2003. Nevertheless, with the present feed ban, fish meal is the only source of PAPs authorized for pig and poultry feed. The decline in the BSE epidemic in the United Kingdom and in most European countries demonstrates that management of the crisis has been, for the most part, successful (Paisley et al., 2008).
5Classical optical microscopy is the only official method for the detection of PAPs in compound feeds or in their ingredients in the European Union (Commission Directive 2003/126/EC). The analysis has two main objectives, which are:
6– the detection of constituents of animal origin,
7– the detection of proteins from terrestrial animals in presence of fishmeal.
8One of the restrictions of classical microscopy is the fact that the method has limited perspectives in terms of species-specific determination of PAPs. However, as stated in the Directive 2003/126/EC, alternative methods can be used to gain more information about the origin of the found PAPs. Different methods have been developed to detect routinely PAPs as well as to identify their origin at the species level and have demonstrated their potential to detect PAPs in feedingstuffs at the benchmark level of 0.1% (w/w) through different studies. However, these methods have not been validated yet at European level through interlaboratory studies. The validation of such efficient and reliable tools is a prerequisite to consider possible lifting of the feed ban for non-ruminants as foreseen in the Commission's TSE Roadmap (European Commission, 2005).
9In 2006, EFPRA (European Fat Processors and Renderers Association) proposed the re-entry of certain PAPs for use in feeds (EFPRA, 2006) respecting the intra species ban laid down in Regulation (EC) n°1774/2002. More recently, EFPRA requested that DG Health and Consumer Protection gives serious consideration to the use of non-ruminant PAPs produced from poultry and porcine sources in feeds for aquatic species (Aqua-feeds) (Woodgate, 2007a). According to EFPRA there are several reasons for such an approach, namely:
10– the availability of non-ruminant PAPs from category 3 ABPs processed in registered plants,
11– PAPs are sustainable and their use in feeds is the most environmentally option,
12– available European PAPs can answer the demand of the Aquafeed market without affecting other markets such as for petfood,
13– there are precedents in Chile and Canada where PAPs are freely used to develop successfully aquafeed diets mainly by the substitution of fish meal with terrestrial non-ruminant proteins.
14Beside the re-entry of non-ruminant proteins in the feed sector, EFPRA called also into question the concept of “zero tolerance” and asked to consider the issue of threshold limits. Zero tolerance means that feedingstuffs containing traces of PAPs other than fish meal cannot be used in animal nutrition, regardless of the corresponding concentration of PAPs in the feedingstuffs. EFPRA recommended a 2% threshold limit for the presence of ruminant PAPs in non-ruminant PAPs as a safe level based on a risk study conducted by Det Norske Veritas Ltd. (DNV) for EFPRA. If accepted whatever the level, the use of tolerance limits would be a new challenge requiring control tools that are also able to quantify accurately the level of PAPs. Tests for the detection of animal constituents in feeds were already reviewed by Momcilovic et al. (2000), Gizzi et al. (2003a) and van Raamsdonk et al. (2007). This review takes also into account the latest developments and studies regarding the quantification issue.
2. Detection of PAPs
2.1. Detection of animal particles
15The classical microscopy. The analytical method for the determination, i.e. detection and identification, of animal constituents in feed as defined in Commission Directive 2003/126/EC entirely relies on the classical microscopy for official controls. The current Directive text results from an in depth revision of Directive 98/88/EC (now repealed). An intercomparison study carried out by the IRMM (Gizzi et al., 2003b) revealed that the different interpretation of the microscopic method as laid down in Directive 98/88/EC resulted in significant differences in sensitivity, specificity and accuracy of the method. The revision was intended to improve the PAPs detection using microscopy by harmonizing the methodology for both qualitative and quantitative analyses. In this section only the qualitative aspect will be considered. The quantitative issue will be discussed in the section 3.
16Practically, the microscopic qualitative determination is realized on different subsamples obtained from the original feed material (or after grinding if needed): the raw material and the concentrated fraction. The concentrated fraction, also referred to as sediment, is obtained through a sedimentation process in tetrachloroethylene that will gather particles above a well-defined density. For this sedimentation, either conical beakers or closed sedimentation funnels can be used. Raw and concentrated materials have to be sieved and the obtained fractions examined by means of compound and stereo microscopes. Various mounting media, like glycerol or paraffin oils, are proposed to the analyst for slide preparation, provided the physicochemical properties of those media allow to maintain the air inside the bone lacunae which facilitates the structure detection by the analyst. The Directive authorizes also the use of different staining reagents such as alizarin red and cystine for enhancing respectively structures such as bones or, fish scales on one hand and hairs and feathers on the other hand.
17Recent collaborative studies (Veys et al., 2007a; 2007b; van Raamsdonk et al., 2008) provided evidence of the global reliability of optical microscopy among control laboratories. Table 1 gives a summary of the overall performances inside some networks of laboratories. Time-evolution performance parameters show that correct detection skills can be improved by continuous training and iterations of proficiency evaluations. In the IAG (International Association for Feedingstuffs Analysis – Section Feedingstuffs Microscopy) 2008 study, the sensitivity expressed in terms of the percentage ratio of correct identification for terrestrial particles by classical microscopy is particularly high for a material adulterated by only 0.05% terrestrial MBM with a value of 95% (from 43 analyses). Moreover, when compared to the two other collaborative studies discussed in this paper, the latter ring test presented a relatively high percentage of false positive results for the presence of fish. It is assumed that at least some of the particles could be misinterpreted and possibly characterized as fish although no direct evidence for this was found.
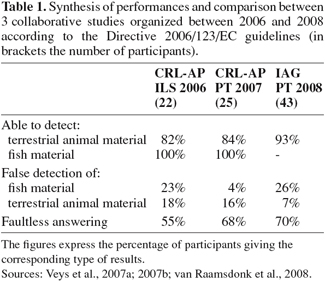
18Reviewing reports and papers concerning classical microscopy indicate the need for further fine-tuning of the 2003/126/EC Directive method. For instance, the initial portion of at least 5 g of sample material to be taken for preparing the different fractions could be fixed to a higher value (Veys et al., 2007a; van Raamsdonk et al., 2008). Another concern is the use of various devices for the sedimentation process and the lack of harmonization of slide preparation which might also be a source of heterogeneous results (Gizzi et al., 2004; van Raamsdonk et al., 2003; 2004; Von Holst et al., 2006). Moreover, the present zero tolerance policy regulating the feed ban is only applicable when a method strictly applied by two operators on a same material is able to yield the same results. Regarding the expression of the results some amendments are also needed. Effectively in cases of very low contamination levels (e.g. < 0.01%) or in cases of natural cross contamination, such as that from rodents or birds, it might be suggested to provide extra information (e.g. number and type of particles detected) in order to highlight authorities on the possible origin of the contamination.
19The near infrared microscopic methods. Near infrared microscopic methods are based on the use of the infrared spectra of individual particles to discriminate the origin of the feed compounds making up the samples. The NIR microscopy (NIRM) method follows exactly the same protocol for sample preparation as classical microscopy. Hundreds of particles from the raw fraction or the sediment fraction are analyzed in order to detect the presence of animal by-products in the sample. Since 1998, the Walloon Agricultural Research Centre (CRA-W) has been pioneer in the development of near infrared microscopic methods based on NIR microscope or NIR imaging systems to detect and quantify meat and bone meal. After several years of development in the framework of national and European projects, the validation of both methods according to international standards has been done. Since 2006, the NIR microscopy and NIR imaging methods are routinely used at CRA-W for routine analysis in the framework of the activities of the Community Reference Laboratory for animal proteins in feedingstuffs (CRL-AP, www.crl.cra.wallonie.be). These analyses are performed under accreditation ISO 17025.
20The first NIRM method using a NIR microscope was developed in 1998 (Piraux et al., 1999; 2000). Later on, within the STRATFEED project (Baeten et al., 2004), the method was significantly improved by:
21– the development of a protocol focusing on the sediment part of the sample which contains mainly denser particles such as bones,
22– the comparison of the performance with classical microscopy,
23– the transfer of the method to another laboratory using a somewhat different instrument but with the help of the discriminant function established at CRA-W (Baeten et al., 2001b; 2004a; 2005c; von Holst et al., 2008). With currently available NIR microscopes the particles are analyzed one by one sequentially and this is a time-consuming process (Baeten et al., 2002). The second microscopic method using NIR imaging system was developed in 2000 and as the former one allows the analysis of the raw and sediment fraction. This system allows the analysis of about 300-500 particles simultaneously and reduces drastically the analytical time (Fernández et al., 2005; Baeten et al., 2005a; 2005c; 2007). These methods have not been validated yet by an interlaboratory study due to the few instruments available. However, there is sufficient in-house validation information available at CRA-W to evaluate the applicability of this technique to the intended purpose.
24The discrimination of terrestrial PAPs from fish by-products can be accomplished by these methods. It has been demonstrated that this discrimination can be done on particles from the sediment fraction, but also on particles from the raw fraction (Baeten et al., 2001a; 2004a; De la Haba et al., 2007a). This is one of the main advantages of the NIRM methods. Discriminant equations that are already available can be used to distinguish the source of the particles in both, raw or the sediment fraction. For the discrimination of the different species of terrestrial animal origin, the results of various studies tend to indicate that the discrimination might be possible. However, because of possible overlapping of the NIR spectra between the different groups, the technique can only give an indication about the origin of the detected PAPs.
2.2. Detection of biological markers
25Animal proteins detected by NIRS. Near infrared spectroscopy (NIRS) is one of the most widely used analytical techniques in the feed sector and is based on absorption of light (absorbance) at selective wavelengths of the electromagnetic spectrum by the organic molecules constituting the analyzed samples. Numerous studies testify the ability of NIR spectroscopy to identify and/or quantify animal ingredients in feed mixtures (Garrido-Varo, 2000; Baeten et al., 2001a; Pérez Martin et al., 2004; Garrido-Varo et al., 2005; Murray et al., 2005; De la Haba et al., 2007b). Murray et al. (2001) showed the potential of NIRS to detect MBM also in fishmeal.
26The major drawbacks of the NIRS technique are that the limit of detection (LOD) is higher than 1% and the method cannot be used alone as legal evidence. Moreover the NIRS can only discriminate the higher taxonomic groups of species (terrestrial animals vs fish). Nevertheless NIRS has a role to play as a first line screening technique in combination with more costly methods to confirm suspect samples.
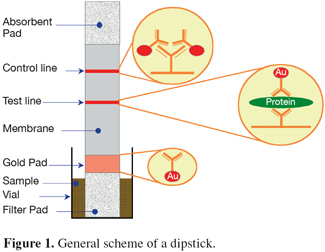
27Animal proteins detected with immunoassays. The principle of the immunochemical techniques is the interaction between the antibody of the test and the antigen in the sample which is in this case a specific processed animal protein. Different designs for the detection of this interaction have been developed but, in the field of the PAPs detection, only the enzyme-linked immunosorbent assay (ELISA) technique and the lateral flow "dipstick" technology have been used. The former method requires the use of typical equipment of an immunoassay laboratory such as a microplate reader whereas dipsticks can be used on-site without specific equipment nor high skilled staff. Figure 1 presents the general scheme of a dipstick.
28Ansfield (1994) worked on an immunoassay using antibodies against thermostable antigens able to withstand severe animal protein rendering process. He developed a patented and in-house validated double sandwich ELISA to detect processed ruminant and porcine proteins in animal compound feeds (Ansfield et al., 2000a; 2000b). The test was subjected to a pre-validation trial conducted by the JRC-IRMM but failed to detect MBM heat treated according to the European Regulation EC/1774/2002 (sterilization with steam pressure ≥ 133°C, 3 bar and 20 min) (van Raamsdonk et al., 2007).
29For a long time, several kits have been developed for the determination of raw or moderately cooked pork in food. Intensive studies demonstrated that the response of the ELISA however was very low when the pork had been heated at the above mentioned sterilization conditions (Hofmann et al., 1995), thus allowing this technique to be used as proof that PAPs containing porcine material have been heat treated according to European legislation. Pallaroni et al. (2001) and von Holst et al. (2001) confirmed these results by investigating the variation of important rendering conditions such as the sterilization temperature or the duration of the treatment on the response of the immunoassays used. More recently, kits specifically devoted to the detection of meat and bone meals are proposed by different companies: the “Reveal for ruminant” tests provided by the American Company Neogen Corporation (Lansing, MI, USA) are lateral flow assays targeting the ruminant heat stable muscle protein Troponin I. Two assays are available and are dedicated to the analysis of different types of samples (feeds and feed ingredients or animal meals). A second dipstick test, “Feedcheck”, developed by Strategic Diagnostics Inc. (SDI – Newark, DE, USA) detects two parameters which are PAPs from all animals and mammalian PAPs. This method uses connective tissue as target and is therefore different from the Neogen test which detects proteins from skeletal muscles. The Reveal for ruminant and Feedcheck tests were subjected to many studies (Fumière et al., 2004; Anonymous, 2004; 2005; Boix et al., 2004; 2006; Klein et al., 2005; Myers et al., 2005). The commercially available test kit developed by Neogen “Reveal for ruminant in feed” passed successfully the ruggedness test of Boix et al. (2004) establishing the impact of various feed ingredients on the analytical results and evaluating the transferability of the method from the laboratory that developed the test to another laboratory. The test showed a sufficient sensitivity at the level of 0.5% ruminant PAPs but insufficient sensitivity when the samples contained 0.1% ruminant PAPs. Some of the blank animal feed samples were wrongly classified as positive. The presence of animal fats from rendering industry might be a source of false positive results especially in pig feeds where this animal fat is frequently used. Some false positive results were also related to beet pulp or citrus pulp used as ingredients in compound feeds. However, these “false” results do not pose any major problem when integrating the method in a global control system, applying the dipstick mainly for screening purposes. Positive samples would then need to be tested by a confirmatory method. The results for the Feedcheck test indicated a good sensitivity of the animal target as almost all positive samples were correctly classified as positive. Only one sample containing PAPs without connective tissue was wrongly classified as negative. With the mammalian target, a large number of false negative results (50%) were observed hinting at a lack of sensitivity and a detection limit above 0.1%. From unpublished results by CRA-W, a cross-reaction of the Feedcheck test with the fishmeals was observed at levels as low as 1.5%. The phenomenon reduces the usefulness of the test as it can give positive results with all feeds containing fishmeal. A study was also published in 2005 by Myers et al. dealing with the performances of the Neogen and SDI tests. The results presented differ slightly from the ruggedness study conducted by IRMM. In the study of Myers et al., the Reveal test demonstrated a perfect selectivity but did not achieved a 0.1% level of sensitivity. The results obtained with the Feedcheck test showed an efficient sensitivity even at a level of 0.1% of MBM but the selectivity was very poor due to the high proportion of false positive results (> 30%). It must be mentioned that the results in this study did not take into account the problems observed elsewhere with ingredients such as beet pulp, citrus pulp and fishmeal.
30Two ELISA kits were also developed by commercial companies: the inhibition ELISA for detecting ruminant PAPs in MBM, feedstuffs and fishmeal proposed by AntibodyShop (Gentofte, Denmark) was successfully implemented in the JRC-IRMM laboratory but the high number of false negative results in samples containing bovine meat and bone meal indicated that the promising performance of the method as shown in the prevalidation study of Boix et al. (2004) could not be confirmed. The method did not appear to be robust enough when transferred to another laboratory. The American company Elisa Technologies Inc. (Gainesville, FL, USA) markets the MELISA-TEK kit which is able to discriminate ruminant and pork Troponin I from other animal troponins and seems to have interesting performances (unpublished data) but was only in-house tested.
31Animal DNA sources detected by polymerase chain reaction. Genetic amplification is presently one of the most efficient ways to detect a well defined DNA target. Among these methods, Polymerase Chain Reaction (PCR) is the most popular and most renowned one. It uses thermal steps to sustain an enzymatic chain reaction that theoretically should double the number of targets at each step. The high forensic value of PCR results is based on the research of specific targets in DNA sequences present in each cell of an organism and conserved at a suitable taxonomic level, commonly at species or groups of animals levels like ruminants or mammals. In that way, different PCR targets (e.g. bovine, ruminant and mammalian targets) can be used to analyze a sample. Nevertheless careful interpretations must be drawn on the results: if positive results obtained with two or more independent PCR tests (different targets) provide converging evidences on the presence of the targeted DNA sources, on the contrary, conflicting results can be due only to different performances (such as sensitivity) of the used tests. Moreover, the PCR approach being a DNA-based technique, detection will only be possible as long as its target molecule is still available, even after the severe sterilization conditions of PAPs as required by European legislation. Therefore, in the particular framework of detection of animal DNA contained in PAPs two important parameters were considered to improve the efficiency of the developed tests:
32– the detection of multi-copy targets instead of single-copy ones: from this point of view, mitochondrial DNA is of major interest as it can be present up to hundred of copies per cell depending on the type of tissue. Different methods were already reported for the identification of different animal species (Krčmář et al., 2003; 2005; Dalmasso et al., 2004; Prado et al., 2004) or of ruminant species (Lahiff et al., 2002; Frezza et al., 2003; Fumière et al., 2006) in feeds using such type of targets. Nevertheless some nuclear targets as short interspersed nuclear elements (SINE) may also be very abundant and were used by Aarts et al. (2006) to detect bovine MBM in animal feed at a level of 0.1%,
33– the detection of short size targets: even if DNA is a rather strong molecule that can survive a lot of drastic processes particularly in bones where DNA is stabilized by mineral sorption (Gotherstrom et al., 2002; Buckley et al., 2008a), rendering will degrade DNA resulting in smaller pieces of DNA. Therefore the target should be small enough (i.e. preferably below 100 bp) to be somewhat below the mean size of the remnant DNA pieces (Fumière et al., 2006) but of course its specificity must be checked (Hird et al., 2006).
34The technique requires an extraction step to isolate the nucleic acids still present in the feed sample. This step is also important for an efficient detection. Indeed, it should be stressed that small DNA fragments are not extracted with the same efficiency as larger template molecules by some DNA extraction systems, particularly for extraction techniques that use DNA precipitation, since small fragments of DNA do not precipitate as easily as large fragments (Hird et al., 2006). The PCR is then performed on a fraction of this extract. During the reaction, a well-defined DNA target, if present, is multiplied several millions of times to make it detectable. The hereby-produced DNA segments are called amplicons and give rise to a fluorescent signal when real time PCR formats are used.
35In an interlaboratory study conducted on behalf of European Commission's DG Health and Consumer Protection (Gizzi et al., 2003b) most of the different PCR techniques failed in terms of required sensitivity and specificity. In fact, only the PCR developed within the STRATFEED project delivered acceptable results, thus supporting the findings of the STRATFEED project about PCR as a potential alternative method for detection of PAPs in feed. At that time, it seemed realistic to consider that samples containing PAPs at 0.5% level (% in weight of MBM of the considered animal species or group of species that the assay can detect) could be detected. It is now established that MBM sterilized at temperatures somewhat above the legal requirement remains detectable at the 0.1% level in feed (% of MBM weight par weight of feed). In the meantime various laboratories improved their PCR techniques to make them applicable to the detection of processed animal proteins at trace level in feed. In a recent JRC-IRMM interlaboratory study from 2006 (Prado et al., 2007) three real time PCR methods were evaluated to determine their applicability for the detection and identification of animal species in feeds. The results indicate that all three laboratories applying their PCR methods were able to detect 0.1% of cattle MBM either alone or in mixtures with different materials such as fishmeal, which demonstrates the high improvements made by this technique, especially when compared with results from former interlaboratory studies.
36Targets were developed for various animal species or groups of species in the STRATFEED project or are described in literature: mammalians, ruminants and cattle are the most common next to other targets such as sheep, pig, chicken, poultry, avian and fish. But a special focus was also given to groups of species such as mice, rats or rodents. Indeed, their detection in feed samples might be useful to explain very low traces of terrestrial animal particles sometimes found in raw material where contamination seems difficult to understand (e.g. beet pulp). Martín et al. (2007) described their method for the specific and qualitative detection of cat, dog, and rat/mouse in food and feedstuffs.
37An important limit of the PCR approach is the fact that animal DNA (belonging to a species or a group of species) detected in a feed sample does not necessarily come from MBM or PAPs. In fact, also allowed feed ingredients such as milk, blood, fat, hydrolyzed proteins produced from ruminant hides and skins or egg products may contain target DNA. The practical impact of this limitation in routine control is not well known yet and may be limited as some of these products are rather expensive so that they are not that widely used in feed. However, in a former study (Bellorini et al., 2005), it was shown that ruminant fat (tallow) could be identified by PCR due to DNA traces present in the residual insoluble impurities (RIIs) of the fat. The identification of tallow by PCR was even possible when the tallow did not contain more than 0.15% RIIs and when the tallow was mixed to porcine fat (lard) at a concentration of 2%. Also in the recent study carried out by the JRC-IRMM (Prado et al., 2007) it was observed that the presence in feed of animal fat such as tallow from the rendering industry might lead to false positive results from a legal point of view when checking for the presence of banned meat and bone meal while analytically the method is correct.
2.3. Combinations of methods
38As stated before, methods based on the amplification of the DNA are a promising solution for the enforcement of the European legislation but, as they detect any source of DNA, positive results can be due to authorized ingredients such as fats or whey powder. NIRM on the other hand can detect meat and bone meal particles in general without being able to assign it to an animal species. In order to eliminate the main drawback of the PCR concerning the positive results due to authorized ingredients, CRA-W develops a strategy combining the NIRM which detects and isolates the particles of MBM origin together with a DNA extraction protocol adapted to a single particle allowing species identification by real time PCR (Fumière et al., 2008). The main challenge was to extract enough DNA able to be amplified by PCR from such a small amount of material. Using a special buffer extracting a DNA ready to be used in a PCR, CRA-W developed a rapid protocol (less than 1 hour) that allows the analysis of the DNA coming from a single PAPs particle with five different targets. Moreover, the NIRM spectra collected from single particle and authenticated by PCR results were used to build species specific spectral databases (Fumière et al., 2005; 2007). The databases are now used to find species-specific spectral markers. The first results obtained show that the strategy used improved also the specificity potential of the NIRM models and allows to give indications about the species origin of the animal particles previously to its PCR analysis. However, some problems of cleaning of the particles need to be solved in order to be absolutely sure that the DNA extracts come only from the particle and not from traces of authorized and target DNA containing feed ingredients attached to the surface of the particle.
39In addition, it has to be mentioned that some other methods like high-performance liquid chromatography (HPLC), electronic nose or even mass spectrometry showed some potential for the detection of PAPs (van Raamsdonk et al., 2007). Recently, Buckley et al. (2008b) described a novel method for the isolation and analysis of the bone collagen (I) α2 chain carboxytelopeptide using the matrix-assisted laser desorption/ionisation-mass spectrometry (MALDI-MS) to distinguish between different species origin.
3. Quantification of PAPs
40Commission Directive 2003/126/EC also contains a procedure to quantify PAPs in feed by classical microscopy. Quantification can only be carried out if bone particles and other identifiable fragments such as fish scales are present in the sediment. Basically, the calculation is computed by using the formula (S x c) / (W x f) x 100, where S is the sediment weight, c (or d in case of fish) is a correction factor for the estimation of the portion of terrestrial bones (or fish bones and scale fragments) in the sediment, W is the weight of the sample material used for the sedimentation and f is a correction factor for the proportion of bones in the constituents of animal origin in the sample examined depending on the type of PAPs present.
41Based on quantification results from 6 laboratories on a set of 10 collection samples, van Raamsdonk et al. (2005) came to the conclusion that the quantification of MBM traces in feed is extremely difficult. This is mainly due to a lack of information on the type of PAPs being detected within a blind sample such as the f that can never securely be estimated. CRL AP ILS 2006 study focused on the implementation and performance evaluation of the quantitative determination of animal constituents in feedingstuffs as described in the 2003/126/EC Directive method (Veys et al., 2007a). The study involved the quantification of the fish meal content of adulterated feed samples at levels ranging from 0.25% to 1.5%. Results of the trial revealed that one third of the participants were unable to apply the method. From the remaining two thirds (i.e. 17 participants) it appeared that aside a global overestimation of the percentage of fish meal, the reproducibility – or interlaboratory variability – was tremendously high (RSDR ranging from 85-116%) although the repeatability – or within-laboratory variability – was nonetheless satisfying (RSDr ranging from 12-30%). Veys et al. (2007a) deduced that the main source of variation was almost likely the d factor and not the sedimentation process (or S and W parameters) nor the f factor. The assumption of d as main cause of variation is supported by two arguments. At first the quantification method from the Directive does not explain how to evaluate the c and d factors and secondly this factor almost entirely depends on the ability of the analyst to discriminate between fish bones and scales – in the CRL-AP ILS 2006 scope – and other particles from the sediment. This hypothesis was confirmed by a second study, referred as CRL-AP ILS 2007, conducted by the same organizers (Veys et al., 2008). The CRL-AP ILS 2007 aimed at validating a calculation tool developed by the CRL-AP for the quantification method in order to avoid computation errors, but also a way of evaluating d according to a clearly defined standard counting procedure (Veys et al., 2008). All participants have applied the same protocol: d was plainly defined and quantification was performed in an harmonized way with respect the use of a counting grid in the eye-piece of the microscope, a mandatory alizarin red staining of the sediment, the number of slides and the number of fields to be observed, the final magnification to be used and a fixed value of 0.10 for f. Regardless of those standardizations, the calculated values were still overestimated. The reproducibility was improved but remains unsatisfying for a reliable quantification (RSDR ranging from 50-84%). The repeatability was however comparable to that from the former CRL-AP ILS 2006, thus satisfying – at least concerning the percentages of adulteration above 0.4% of fish.
42Regarding the quantification of PAP by NIRM methods, several studies have briefly shown their potential as well for the sediment as for the raw fraction (Baeten et al., 2004; 2005b; Fernández Pierna et al., 2005). NIRM and NIR imaging have a potential to quantify PAP in raw fraction of the compound feeds. Nevertheless, the existing protocols (mainly developed for qualitative analysis) need further developments in order to include mandatory steps for the quantification aspect.
43With real time PCR, the amplification of the DNA target can be followed during the reaction itself (generally on-line but not necessarily) and gives information on the kinetics of the reaction. Therefore it can be used for quantization purposes. However it should be stressed that basically the technique quantifies a number of copies of targets. In the case of PAP detection in feed, it is difficult to use this parameter because there is no straight relationship between the weight of PAP and its content in number of copies of a defined target. Indeed the number of targets available can be material-dependent (e.g. the number of mitochondrial targets available is largely tissue-dependent) but in addition, the rendering process itself has a huge effect on the number of remaining exploitable targets. The impact of sterilization has been clearly evidenced with meat and bone meal processed in a batch-type commercial rendering plant at different temperatures up to 141°C (Chiappini et al., 2005; Fumière et al., 2006). At that temperature DNA is still detectable from the pure MBM but the absolute number of copies of the target decreases largely. Special treatments with acids or bases, used for instance during extraction of gelatin, can also have a great damaging effect on DNA although it seems that the DNA in bone particles is much better protected (e.g. in fossils DNA has been kept for very long periods – Buckley et al., 2008a). On the contrary, the presence of authorized ingredients containing many intact targets such as whey would lead to an over-estimation of the PAP content. Nevertheless, different authors already attempted to use the technique to quantify the level of PAP present in a feedingstuff (Fumière et al., 2006; Frezza et al., 2008) but they pointed out that it was mainly a demonstration of the feasibility on a specific set of samples while for routine applications it is not possible to use this technique on unknown samples.
44The response of immunochemical tests can be strongly influenced by the tissue content and/or different process parameters (temperature, pressure, time). In the case of the Reveal kits, an optical reading of the test can be performed with the Accuscan Reader provided by Neogen Corporation and the intensity of the signal can be quantified on a scale from 0 to 4. The automated reading of the test allows an objective conclusion independent of the user especially with samples giving a very faint positive signal. The read parameter is however unfit for quantifying the PAP content. Nevertheless, within the safe re-entry of non-ruminant PAPs in feeds for aquatic species prospect, the European Fat Processors and Renderers Association (EFPRA) proposed a 2% tolerance level of contamination of non-ruminant PAP by ruminant PAP as it would have negligible impact on TSE risks. The use of the Reveal for ruminant in feed test as the tool for a semi-quantitative analysis of PAPs in combination with other methods was therefore recommended by EFPRA (Woodgate, 2007b). But, in the case of the Reveal for ruminant kit, some disruptive effects (e.g. masking of ruminant material with pig PAPs) can also occur (Fumière et al., 2004) and a full evaluation of the exact potential of the test has to be conducted.
4. Discussion and conclusion
45Up to now, none of the techniques considered enable a full implementation of the European Legislation (detection at low level, identification at species level and quantification of PAPs) in order to allow a reappraisal of the total feed ban:
46– The classical microscopy is mainly based on the detection of bones. With the sedimentation step, the LOD of the technique is very low. Nevertheless, the determination of the species origin is limited and the quantification requires the use of factors introducing sources of errors.
47– The NIRM technology has characteristics to be used as a screening method: it does not need experienced staff and can be automated. It is also a technique that can be used in addition to or in combination with other methods. It has also the potential to work on the raw fraction as well as on the sediment. Nevertheless, the equipment required for it is expensive and not yet largely used.
48– Powerful PCR methods have been developed and real time PCR allows to give after suitable transformation a quantitative result. But in order to obtain an indispensable sensitivity difficult to reach with such processed materials, all the efficient methods target markers present in multiple copies in cells. The number of copies of this type of targets is often tissue dependent and does not allow a relative quantification as it is possible with genetically modified organism (GMO). The only scientifically sound way to express quantitatively the results would be to calculate the number of PCR amplifiable targets present in a sample. For that purpose, a calibration method based on the use of plasmids is under development at CRA-W (unpublished results) within the European SAFEED-PAP project. This tool would also allow an efficient transferability from a platform (thermocycler + PCR reagents) to another one, especially with respect to the definition of cut-off thresholds for the determination of positive or negative PCR results. The procedure would have the advantage to take into account the different parameters able to influence the efficiency of the PCR and to make possible an easy and standardized use of the technology by any laboratory. Concerning the use of authorized ingredients which can be sources of target DNA, the problem remains unresolved for the PCR. Its combination with the NIRM could be a sophisticated solution to evaluate.
49– Due to their high throughput, immunoassays are still considered as good candidates to be screening methods incorporated within a global control system but some requirements such as a higher sensitivity and a better specificity need to be fulfilled. The European SAFEED-PAP project works on these issues and a special effort is done on the improvement of the extraction procedure. However, immunoassays will remain indicative methods needing to be confirmed by other methods with forensic value like PCR.
50A possible solution would be an analytical system using and combining the methods according to their potential to answer to the following questions:
51– Does the sample contain PAPs?
52– What is the species origin of the PAPs present in the sample?
53– If animal products are present, do they come from an authorized or a forbidden ingredient?
54– What is the level of PAPs content?
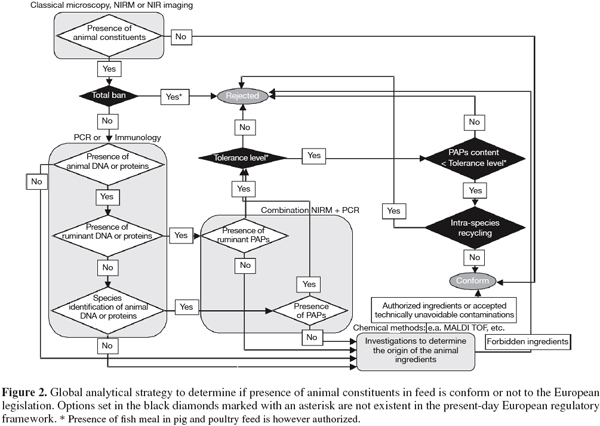
55– According to the answers to the previous questions, it can be established whether the feed is in compliance with the legislation. The figure 2 suggests a possible decision tree taking into account possible changes of the legislation (intra-species ban and tolerance level). Due to its high detection efficiency, classical microscopy will probably remain a first line control tool but additional methods will be unavoidable to allow the full implementation of the legislation.
Bibliographie
Aarts H.J.M. et al., 2006. Detection of bovine meat and bone meal in animal feed at a level of 0.1%. J. AOAC Int., 89(6), 1443-1446.
Anonymous, 2004. AOAC Research Institute. Certificate of Performance TestedSM Status. Certificate n°010405, http://www.aoac.org/testkits/2008_010405_certificate.pdf, (28.11.08).
Anonymous, 2005. AOAC Research Institute. Certificate of Performance TestedSM Status. Certificate n° 070501, http://www.aoac.org/testkits/2008_070501_certificate.pdf, (28.11.08).
Ansfield M., 1994. Production of a sensitive immunoassay for detection of ruminant proteins in rendered animal material heated to > 130°C. Food Agric. Immunol., 6, 419-433.
Ansfield M., Reaney S.D. & Jackman R., 2000a. Performance assessment and validation of a sensitive immunoassay for detection of ruminant and porcine heat stable proteins in compound animal feedstuffs. Food Agric. Immunol., 12, 285-297.
Ansfield M., Reaney S.D. & Jackman R., 2000b. Production of a sensitive immunoassay for detection of ruminant and porcine proteins, heated to > 130°C at 2.7 bar, in compound animal feedstuffs. Food Agric. Immunol., 12, 273-284.
Baeten V. & Dardenne P., 2001a. The contribution of near infrared spectroscopy to the fight against the mad cow epidemic. NIRS news, 12(6), 12-13.
Baeten V., Michotte Renier A., Sinnaeve G. & Dardenne P., 2001b. Analysis of feedingstuffs by near-infrared microscopy (NIRM): detection and quantification of meat and bone meal (MBM). In: Proceedings of the 6th International symposium on food authenticity and safety FASIS conference, November 28-30, 2001, Nantes, France. Nantes, France: Eurofins Scientific, 1-11.
Baeten V. & Dardenne P., 2002. Spectroscopy: developments in instrumentation and analysis. Grasas y aceites, 53(1), 45-63.
Baeten V. et al., 2004. The near infrared microscopic (NIRM) method: combination of the advantages of optical microscopy and near-infrared spectroscopy (WP5-NIRM). In: Strategies and methods to detect and quantify mammalian tissues in feedingstuffs. Brussels: European Commission.
Baeten V. & Dardenne P., 2005a. Applications of near-infrared imaging for monitoring agricultural food and feed products. In: Bhargava R. & Levin I.W., eds. Spectrochemical analysis using infrared multichannel detectors. Oxford, UK: Blackwell Publishing, 283-302.
Baeten V. et al., 2005b. Detection of banned meat and bone meal in feedingstuffs by near-infrared microscopy analysis of the sediment fraction. Anal. Bioanalytical Chem., 382, 149-157.
Baeten V., Fernández Pierna J.A., Michotte Renier A. & Dardenne P., 2005c. Imagerie infrarouge : analyse des aliments à destination animale. Techn. Ing., RE 34, 1-8.
Baeten V., Fernández Pierna J.A. & Dardenne P., 2007. Hyperspectral imaging techniques: an attractive solution for the analysis of biological and agricultural materials. In: Grahn H.F. & Geladi P., eds. Techniques and applications of hyperspectral image analysis. Chichester, UK: Wiley, 289-312.
Bellorini S. et al., 2005. Discriminating animal fats and their origins: assessing the potentials of Fourier transform infrared microscopy, gas chromatography, immunoassay and polymerase chain reaction techniques. Anal. Bioanalytical Chem., 382, 1073-1083.
Boix A. et al., 2004. Determination of Processed Animal Proteins (PAPs) including meat and bone meal (MBM) in feed. Part I: Intercomparison study for the determination of PAPs in feed using microscopy. Part II: Prevalidation study for the detection of PAPs in feed by immunoassays. Geel, The Netherlands: JRC-IRMM, http://irmm.jrc.ec.europa.eu/html/activities/meat_and_bone_meal/PAP_MBM_2004_report.pdf, (26.11.08).
Boix A., Serano F., Bellorini S. & von Holst C., 2006. Ruggedness study of immunoassays for processed animal proteins detection in feed: Neogen Reveal for Ruminant Feed Test System. Geel, The Netherlands: JRC-IRMM, http://irmm.jrc.ec.europa.eu/html/activities/meat_and_bone_meal/Ruggedness_study_september06/pdf, (26.11.08).
Buckley M. et al., 2008a. Comparing the survival of osteocalcin and mtDNA in archaeological bone from four European sites. J. Archeol. Sci., 35, 1756-1764.
Buckley M., Collins M. & Thomas-Oates J., 2008b. A method of isolating the collagen (I) alpha 2 chain carboxytelopeptide for species identification in bone fragments. Anal. Biochem., 374(2), 325-334.
Chiappini B. et al., 2005. A real time PCR approach for ruminant-specific DNA quantification indicates a correlation between DNA amount and MBM heat treatments. J. AOAC Int., 88(5), 1399-1403.
Commission Directive 98/88/EC of 13 November 1998 establishing guidelines for the microscopic identification and estimation of constituents of animal origin for the official control of feedingstuffs. Off. J. Eur. Communities, L318, 27.11.98, 45-50.
Commission Directive 2003/126/EC of 23 December 2003 on the analytical method for the determination of constituents of animal origin for the official control of feedingstuffs. Off. J. Eur. Union, L339, 24.12.2003, 78-84.
Commission Regulation (EC) n°1234/2003 of 10 July 2003 amending Annexes I, IV and XI to Regulation (EC) n°999/2001 of the European Parliament and of the Council and Regulation (EC) n°1326/2001 as regards transmissible spongiform encephalopathies and animal feeding. Off. J. Eur. Union, L173, 11.7.2003, 6-14.
Dalmasso A. et al., 2004. A multiplex PCR assay for the identification of animal species in feedstuffs. Mol. Cell. Probes, 18, 81-87.
De la Haba M.J. et al., 2007a. Discrimination of fish bones from other animal bones in the sedimented fraction of compound feeds by near infrared microscopy (NIRM). J. NIRS, 15, 81-88.
De la Haba M.J., Garrido-Varo A., Pérez-Marín D.C. & Guerrero J.E., 2007b. Near infrared analysis as a first-line screening technique for identifying animal species in rendered animal by-products meals. J. Near Infrared Spectrosc., 15, 237-245.
EFPRA, 2006. EF/06/108 proposal for re-entry of certain PAPs for use in feeds: a discussion document, 9 October 2006. Brussels: EFPRA.
European Commission, 2005. The TSE Roadmap, http://ec.europa.eu/food/food/biosafety/bse/roadmap_en.pdf, (26.11.08).
Fernández Pierna J.A. et al., 2005. Combination of SVM and NIR imaging spectroscopy for the detection of MBM in compound feeds. J. Chem., 18(7-8), 341-349.
Frezza D. et al., 2003. A competitive polymerase chain reaction-based approach for the identification and semiquantification of mitochondrial DNA in differently heat-treated bovine meat and bone meal. J. Food Protect., 66, 103-109.
Frezza D. et al., 2008. Standard and light-cycler PCR methods for animal feedstuffs. Innovative Food Sci. Emerging Technol., 9, 18-23.
Fumière O., Osmanaj I., Lafortune M.-G. & Berben G., 2004. Rapid detection of processed animal protein in feed with ReVeal® Ruminant strip test. In: Proceedings of Rapid Methods Europe 2004, March 25-26, 2004, Noordwijk aan Zee, The Netherlands. Bilthoven, The Netherlands: Rapid Methods Europe.
Fumière O. et al., 2005. Speciation of mammalian meat and bone meal particles with near infrared microscopy and real time polymerase chain reaction. In: Burling-Claridge G.R., Holroyd S.E. & Sumner R.M.W., eds. Proceedings of the 12th International conference on near infrared spectroscopy, Auckland, New Zealand, April 9-15, 2005. Hamilton, New Zealand: New Zealand Infrared Spectroscopy Society Incorporated, 129-132.
Fumière O. et al., 2006. Effective PCR detection of animal species in highly processed animal by-products and compound feeds. Anal. Bioanalytical Chem., 385, 1045-1054.
Fumière O. et al., 2007. Original combination of real time PCR and NIRM for the detection and the speciation of animal particles. In: Feed Safety International Conference 2007: methods and challenges, November 27-28, 2007, Namur, Belgium, http://safeedpap.feedsafety.org/fs2007/posters/fumiere.php, (19.11.08).
Fumière O. et al., 2008. Development of an original DNA extraction protocol for the species specific identification of PAPs particles in feeds using NIRM and Real Time PCR. In: Proceedings of Rapid Methods Europe 2008, January 21-23, 2008, Noordwijkerhout, The Netherlands. Bilthoven, The Netherlands: Rapid Methods Europe.
Garrido-Varo A., 2000. La spectroscopie proche infrarouge : une technologie d'appui pour un service intégral en alimentation animale. In: Bertrand D. & Dufour E., eds. La spectroscopie infrarouge et ses applications analytiques. Paris : Editions Tec & Doc-Lavoisier, 273.
Garrido-Varo A. et al., 2005. Near infrared spectroscopy for enforcement of European legislation concerning the use of animal by-products in animal feeds. Biotechnol. Agron. Soc. Environ., 9(1), 3-9.
Gizzi G. et al., 2003a. An overview of tests for animal tissues in feeds applied in response to public health concerns regarding bovine spongiform encephalopathy. Rev. Sci. Tech. Off. Int. Epizoot., 22(1), 311-331.
Gizzi G. et al., 2003b. Intercomparison study for the determination of processed animal proteins including meat and bone meal in animal feed. Final report of the Administrative arrangement n°B5-1000/02/000483. Geel, Belgium: JRC-IRMM, 100.
Gizzi G. et al., 2004. Determination of processed animal proteins, including meat and bone meal in animal feed. J. AOAC Int., 87(6), 1334-1341.
Götherström A., Collins M.J., Angerbjorn A. & Liden K., 2002. Bone preservation and DNA amplification. Archaeometry, 44, 395-404.
Hird H. et al., 2006. Effect of heat and pressure processing on DNA fragmentation and implications for the detection of meat using real time polymerase chain reaction. Food Addit. Contaminants, 23(7), 645-650.
Hofmann K., Fischer K., Müller E. & Kemper V., 1995. Versuche zum Nachweis der Erhitzungseffektivität bei Fleischkonserven und Tiermehlen. Fleichwirtschaft, 75(10), 1227-1231.
Klein F., Lupo T., Pielack D. & Mozola M., 2005. Validation study of a lateral-flow immunoassay for detection of ruminant by-product material in animal feeds and feed ingredients. J. AOAC Int., 88(6), 1583-1592.
Krčmář P. & Renčová E., 2003. Identification of species-specific DNA in feedstuffs. J. Agric. Food Chem., 51, 7655-7658.
Krčmář P. & Renčová E., 2005. Quantitative detection of species-specific DNA in feedstuffs and fish meals. J. Food Protect., 68(6), 1217-1221.
Lahiff S. et al., 2002. Real time polymerase chain reaction detection of bovine DNA in meat and bone meal samples. J. Food Protect., 65, 1158-1165.
Martín I. et al., 2007. Detection of cat, dog, and rat/mouse tissues in food and animal feed using species-specific polymerase chain reaction. J. Anim. Sci., 85, 2734-2739.
Momcilovic D. & Rasooly A., 2000. Detection and analysis of animal material in food and feed. J. Food Prot., 63, 1602-1609.
Murray I., Aucott L.S. & Pike I.H., 2001. Use of discriminant analysis on visible and near infrared reflectance spectra to detect adulteration of fish meal with meat-and-bone meal. J. Near Infrared Spectrosc., 9, 297-311.
Murray I. et al., 2005. Macroscopic near-infrared reflectance spectroscopy. In: EUR 21124 Strategies and methods to detect and quantify mammalian tissues in feedingstuffs. Luxembourg: European Commission, 156.
Myers M. et al., 2005. Evaluation of two commercial lateral-flow test kits for detection of animal proteins in animal feed. J. Food Protect., 68(12), 2656-2664.
Paisley L.G. et al., 2008. Risk analysis of transmissible spongiform encephalopathies in animals: state-of-the-art. Int. J. Risk Assess. Manage., 8(3), 214-242.
Pallaroni L., Björklund E., von Holst C. & Unglaub W., 2001. Determination of rendering plant sterilisation conditions using a commercially available ELISA test kit developed for detection of cooked beef. J. AOAC Int., 84(6), 1844-1890.
Pérez-Martin M.D. et al., 2004. Detection and quantification of mammalian meat and bone meal in compound feedingstuffs using NIR. In: Davies A.M.C. & Garrido-Varo A., eds. Proceedings of the 11th International conference on near infrared spectroscopy, April 6-11, 2003, Cordoba, Spain. Chichester, UK: NIR Publications, 667-671.
Piraux F. & Dardenne P., 1999. Feed authentication by near-infrared microscopy. In: Davies A.M.C. & Giangiacomo R., eds. Proceedings of the 9th international conference on near-infrared microscopy, June 13-18, 1999, Verona, Italy. Chichester, UK: NIR Publications.
Piraux F. & Dardenne P., 2000. Microscopie-NIR appliquée aux aliments du bétail. Biotechnol. Agron. Soc. Environ., 4(4), 226-232.
Prado M. et al., 2004. Application of a polymerase chain reaction (PCR) method as a complementary tool to microscopic analysis for the detection of bones and other animal tissues in home-made animal meals. J. Sci. Food Agric., 84, 505-512.
Prado M. et al., 2007. Detection of ruminant meat and bone meals in animal feed by real time Polymerase Chain Reaction: result of an interlaboratory study. J. Agric. Food Chem., 55, 7495-7501.
Regulation (EC) n°999/2001 of the European Parliament and of the Council of 22 May 2001 laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies. Off. J. Eur. Communities, L147, 31.05.01, 1-40.
Regulation (EC) n°1774/2002 of the European Parliament and of the Council of 3 October 2002 laying down health rules concerning animal by-products not intended for human consumption. Off. J. Eur. Communities, L273, 10.10.2002, 1-95.
SAfe FEED. Processed animal proteins detection of presence of species-specific processed animal proteins in animal feed, http://safeedpap.feedsafety.org, (17.11.08).
van Raamsdonk L.W.D. & van der Voet H., 2003. A ring trial for the detection of animal tissues in feeds in the presence of fish meal. Report. Wageningen, The Netherlands: RIKILT.
van Raamsdonk L.W.D. et al., 2004. The microscopic detection of animal proteins in feeds. Biotechnol. Agron. Soc. Environ., 8(4), 241-247.
van Raamsdonk L.W.D. et al., 2005. Microscopic detection of animal by-products in feed. In: EUR 21124 Strategies and methods to detect and quantify mammalian tissues in feedingstuffs. Luxembourg: European Commission, 156.
van Raamsdonk L.W.D. et al., 2007. New developments in the detection and identification of processed animal proteins in feeds. Anim. Feed Sci. Technol., 133, 63-83.
van Raamsdonk L.W.D. et al., 2008. The 2008 Dutch NRL/IAG proficiency test for detection of animal proteins in feed. Wageningen, The Netherlands: RIKILT, http://library.wur.nl/way/bestanden/clc/1876397.pdf, (20.11.08).
Veys P. & Baeten V., 2007a. CRL-AP Interlaboratory study 2006 final report. Gembloux, Belgium: CRA-W, http://ec.europa.eu/food/food/biosafety/resources/interlaboratory2006_en.pdf, (18.11.08).
Veys P. & Baeten V., 2007b. CRL-AP Proficiency test 2007 final report. Gembloux, Belgium: CRA-W.
Veys P. & Baeten V., 2008. CRL-AP Interlaboratory study 2007 final report. Gembloux, Belgium: CRA-W.
von Holst C., Unglaub W. & Anklam E., 2001. Post process product control of rendering plant sterilization conditions by ELISA. J. AOAC Int., 84(6), 1793-1799.
von Holst C. et al., 2006. Determination of processed animal proteins in feed: the performance characteristics of classical microscopy and immunoassay method. Food Addit. Contaminants, 23(3), 252-264.
von Holst C. et al., 2008. Transferability study of a near-infrared microscopic method for the detection of banned meat and bone meal in feedingstuffs. Anal. Bioanalytical Chem., 392, 313-317.
Woodgate S.L., 2007a. EF/07/85 Proposal for the approval of non-ruminant processed animal proteins to be used in feeds for aquatic species (Aqua-feeds), October 18, 2007. Brussels: EFPRA.
Woodgate S.L., 2007b. Feed chain: specificity and challenges. Feed safety international conference 2007: methods and challenges, November 27-28, 2007, http://safeedpap.feedsafety.org/fs2007/lectures/woodgate.php, (19.11.08).
Pour citer cet article
A propos de : Olivier Fumière
Centre wallon de Recherches agronomiques (CRA-W). Département Qualité des Productions agricoles. Chaussée de Namur, 24. B-5030 Gembloux (Belgique). E-mail: fumiere@cra.wallonie.be
A propos de : Pascal Veys
Centre wallon de Recherches agronomiques (CRA-W). Département Qualité des Productions agricoles. Chaussée de Namur, 24. B-5030 Gembloux (Belgique).
A propos de : Ana Boix
European Commission. Joint Research Centre – Institute for Reference Materials and Measurements (JRC – IRMM). Retieseweg, 111. B-2440 Geel (Belgium).
A propos de : Christoph von Holst
European Commission. Joint Research Centre – Institute for Reference Materials and Measurements (JRC – IRMM). Retieseweg, 111. B-2440 Geel (Belgium).
A propos de : Vincent Baeten
Centre wallon de Recherches agronomiques (CRA-W). Département Qualité des Productions agricoles. Chaussée de Namur, 24. B-5030 Gembloux (Belgique).
A propos de : Gilbert Berben
Centre wallon de Recherches agronomiques (CRA-W). Département Qualité des Productions agricoles. Chaussée de Namur, 24. B-5030 Gembloux (Belgique).






