- Startpagina tijdschrift
- volume 13 (2009)
- numéro 1
- Genotype by environment interaction in dairy cattle
Weergave(s): 0 (0 ULiège)
Download(s): 0 (0 ULiège)
Genotype by environment interaction in dairy cattle

Nota's van de redactie
Received on January 10, 2008, accepted on May 6, 2008
Résumé
Interactions entre génotype et environnement chez les bovins laitiers. Cette revue bibliographique a permis d’identifier la présence de G × E chez les bovins laitiers à partir des performances phénotypiques enregistrées dans différents environnements. Les méthodes utilisées pour l’investigation de G × E ont été discutées. L’importance et l’échelle de grandeur de ces interactions basées sur l’utilisation de ces méthodes sont signalées. L’existence de G × E est essentiellement confirmée en présence de grandes différences entre les environnements de production et/ou de distances génétiques entre les génotypes. Les effets environnementaux ont été agrégés dans les différentes études avec une identification assez synthétique des composants du milieu, excepté quelques travaux récents utilisant une définition plus fine de l’environnement. Les implications de G × E sur les programmes de sélection seront discutées. Les éleveurs devraient sélectionner les génotypes dans les conditions environnementales dans lesquelles ces candidats reproducteurs seront élevés en utilisant un index de sélection combinant les caractères de production et les principaux autres caractères économiques.
Abstract
The aim of this literature review was to identify the existence and scope of genotype by environment interaction (G × E) from reports on dairy cattle populations in different management systems. Methods applied to deal with G × E (controlled experiments and large data modeling) were discussed. A G × E was confirmed essentially when high differences between production environments and/or genotypes (genetically distant genotypes) were observed. Environmental effects were aggregated in most studies and identification of the components of the environment was largely unresolved, with only a few studies based on more definite-descriptors of environment. The implications of G × E on breeding decisions are discussed. Breeders should select genotypes on production traits within environmental conditions comparable to where candidate animals are intended to perform.
Inhoudstafel
1. Introduction
1Milk production will need to nearly double in the world over the next decade to follow population and income growth. The strongest demand for milk and milk products are anticipated for developing countries where an important population growth is expected (Tollens et al., 2004). In addition, internationalization and world globalization will lead to an even freer world dairy market and an enlargement of germplasm exchange in the world. This situation would be translated by an increased intensification and industrialization of production systems and will consequently have profound implications on production systems and the environment. However, the sustainable intensification requires appropriate use of genetic resources with an understanding of the limitations and opportunities of the production environment in which the animals will be maintained. The ability of farmers to respond to environmental conditions such as climate, feed base, food security, and consumer preferences should guarantee a sustainable livestock development.
2In recent decades, dairy cattle breeding has become an increasingly international business and a substantial exchange of Holstein semen has taken place worldwide (Powell et al., 1994). On the other hand, performances of daughters of AI bulls are recorded in various environments in the world. Selection of superior animals, chosen on breeding values from national evaluations, has been operating within countries. However, Banos et al. (1991) reported that across country selection is more profitable under the globalization of dairy industries. Currently, the multiple-trait across country evaluation (MACE) procedure (Schaeffer, 1994) is used by Interbull. This routine evaluation incorporates information on daughters of bulls from different Interbull country members. Then, genetic correlations between countries are estimated from common bulls and three-quarter sibs that have progeny in multiple countries. The international bull breeding values can then be converted to national scales. Thus, using the international evaluation, foreign bulls can be reliably selected for national use. Lohuis et al. (1998) estimated that the global selection can increase rates of genetic response by up to 17% compared to within-country selection.
3In the absence of genotype by environment interaction (G × E), the expected genetic correlation across environments is one. Cooper et al. (1994) reported that only when the genetic correlation among environments is less than one does the G × E impede response to selection. With the current international genetic evaluation of bulls (Interbull), the national trait measures are viewed as different traits depending on the location of herds and using the country member borders as the criterion for differing among environments. Such procedures are ignoring the differences between herds in the same country especially in large countries. It also ignores the similarities between many herds within and between countries (Weigel et al., 2000; Fikse et al., 2003; Zwald et al., 2003a).
4Tropical and developing countries often rely on exotic germplasm for breeding purposes. They however have climatic conditions, production systems, and markets different from those where animals were evaluated. Thus, the G × E can cause a reduced efficiency of their genetic improvement programs. The investigation of G × E in order to thwart this fact was limited and concerned mainly large populations in the northern hemisphere and in a few tropical countries.
5The objective of this paper was to review methods used to study G × E, to asses the importance of G × E, and to determine its effect on the efficiency on selection programs.
2. Genotype by environment interaction: definition and theory
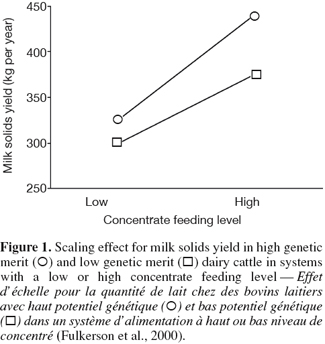
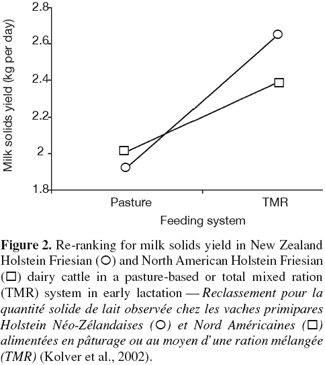
6G × E occurs when performances of different genotypes are not equally affected by different environments (Falconer, 1952). The ability of living organisms (plants or animals) to alter the phenotype in response to changes in the environment is known as phenotypic plasticity or environmental sensitivity (Falconer et al., 1996). When the same genotypes develop different phenotypes in different environments, then there is G × E. When the differences between genotypes vary between environments without changes in their ranking there is scaling effect (Figure 1). However, if the genotypes rank differently in different environment, the effect of G × E is re-ranking of individuals (Figure 2). G × E is of less importance if only scaling effect is obtained because the best selected individuals in one environment would still perform the best in other environments.
7The choice of environment or genotype characterization depends on the aim of each study. Genotype can refer to a genotypic unit (breeds, crossbreds, individuals), but also to a genotypic value (individuals with certain phenotypic or genotypic performances, QTLs, genes). In the same way, environments could be defined as a unit (herd, region, country, etc.), but also as a continuous value (temperature, rainfall, concentrate, feeding level, etc.). Lin et al. (2002) reported that genotype could be classified into three levels in combination with the environment:
8– breed by environment interaction (between-breed interaction),
9– individual by environment interaction (within-breed interaction),
10– gene by environment interaction (within-individual interaction).
11The usual elementary unit for definition of environment in dairy cattle is the herd. Using individual characteristics of each herd as a different environment will lead to great difficulties in comparing different environments. Grouping herds according to their environmental similarities can be an alternative, but availability and accuracy of G × E determinism will depend on the “robustness” of the criterion used for their clustering. In the literature, these characteristics varied from a global, specific, to more detailed definition. Environments have been defined both as between countries with large climatic differences (Stanton et al., 1991; Cienfuegos-Rivas et al., 1999; Costa et al., 2000; Rekaya et al., 2001) as well as within country (Carabaño et al., 1990; König et al., 2005; Gernand et al., 2007). Specific characteristics that have been examined include average herd level, herd size, feeding systems and levels, management, and housing systems (Hill et al., 1983; Cromie et al., 1998; Pryce et al., 1999; Boettcher et al., 2003; Fatehi et al., 2003; Hayes et al., 2003; Nauta et al., 2006). A more limited herd environment characterization based on fine-definite environment descriptors applying canonical correspondence analysis, factor analysis, or principal component analysis, were recently introduced. This approach allows clustering of herds based on fine-definite farm characteristics and might be more efficient and realistic (Weigel et al., 2001; Zwald et al., 2003b; Windig et al., 2005; Haskell et al., 2007).
12The relationship between variations of the phenotypic expression of a genotype under continuous value of the environment is often shown as a reaction norm (Kolmodin et al., 2002). In that case, the phenotypic expression of a genotype is viewed as a function of an environmental parameter (temperature, concentrate). If phenotypes change gradually or continuously over an environment gradient, the reaction approach is appropriate (de Jong, 1995). Plastic genotypes are known by highly variable phenotypes across environments, whereas robust or stable genotypes are known by relatively constant phenotypes across environments (de Jong et al., 2002). When genotypes have significant differences between the quantitative measures of the phenotypic plasticity, then there is a G × E interaction. Differences in the phenotypic plasticity could be explained by the fact that some alleles may only be expressed in some specific environment. Favorable genes in some environments may become unfavorable under other environment conditions. Via et al. (1995) recognize that gene regulation may change depending on the environment.
3. Measures of genotype by environment interaction
13To study G × E, records on both the genotype and the environment are required. The performance of a genotype (i.e. cow) cannot be recorded simultaneously in more than one environment (i.e. countries or regions). Because of the extensive use of AI in the dairy industries, daughters of the same sire are spread in different herds around the world. Nevertheless, performances are obtained according to milk recording schemes based on “universal” guidelines. Information on the environment in which the record was taken are still less “detailed”. To overcome the lack of detailed information obtained from routine milk recording data, G × E measure can be based on experiments. A compromise between costs, availability of data, and experimental unit scale should be taken into account.
3.1. Controlled experiments
14The most reported experimental studies investigating the existence of G × E in dairy cattle have taken place in experimental farms in The Netherlands, Australia, Ireland, and New Zealand. The genotype was generally defined as a different strain of Holstein-Friesian and compromised specific groups based on the level of genetic merit. Environments were usually defined based on differences in feeding level and system. Experimental designs and protocols involved hundreds of animals. Published results on the lack or existence of G × E concerned milk production (Veerkamp et al., 1995; Kolver et al., 2002; Beerda et al., 2007), body score condition (Veerkamp et al., 1994; Horan et al., 2005; McCarthy et al., 2007a), body dimensions, body weights and puberty (Macdonald et al., 2007), health, fertility (Pryce et al., 1999; Ouweltjes et al., 2007), and energy balance (Berry et al., 2007; Beerda et al., 2007). Most of these studies used differences in coefficients from regressing phenotypic performances on environments as indicators of G × E. In general, using experimental herds is expensive but more illustrative. On the other hand, results ought to be viewed as representing a genetic group “strain” and should be cautiously extrapolated out to the general population.
3.2. Modeling genetic variation
15There are three main methods used for estimating G × E: interaction model, character state model, and reaction norm model. These models can be viewed as an extension of the simple and traditional genetic model for quantitative traits in which the phenotype (P) is considered as the sum of only independent genetic (G) and environmental effects (E) [P = G + E] (Falconer et al., 1996).
16The interaction model represents an extension of the traditional genetic model by an inclusion of the random interaction of genotype and environment and thus P = G + E + G × E. The most famous application is the use of the sire x herd interaction (Van der Werf et al., 1991; Dimov et al., 1995). With unbalanced data and in the presence of heterogeneity of variances among environments (herds), the interaction interpretation is difficult when applying this model (Dickerson, 1962). The sire x herd method permits a global description of the effect without allowing individual variations. In addition, the genetic additive relationships among sires are not considered in the estimation of G × E which is only an additional environmental effect.
17Genes may show different expressions under different environments. Falconer (1952) described the expression of a trait in different environments as different characters, or "character states". Thus, the performance of animals in different environments should be regarded as separate traits. Therefore, with the character state model, the genetic correlation between the same types of performance but measured in different environments is used to measure the G × E. The animal breeding analogy with the character state model is the multi-trait model, where performances in different environments are regarded as different and genetically correlated traits. With the character state model, the total additive genetic variance of the plastic trait among environments can be partitioned into the genetic variances of the character states within each environment and the genetic covariance between environments. The later is related to the G × E variance (de Jong et al., 2002). When only two environments are studied, a bivariate analysis is the method for estimating genetic correlation. If more than two environments are involved, a multivariate analysis is applied and genetic correlation can be estimated for any pairs of environment combinations. A well-known application of the character state model is the estimation of bulls' breeding values by Interbull using the Multiple Across Country Evaluation (MACE). Each country is considered as a different environment and the correlation between all pairs of country combinations is estimated. The character state model is known by the flexibility of its variance-covariance structure. However, the number of environments should be kept limited to meet computation requirements and convergence limitations. Moreover, this model is restricted to discrete environments and requires their classification into groups. The accuracy of correlation estimates between environments depends on the best representation of these groups. Clustering methods can be used to group environments with reference to major environments descriptors (e.g. Weigel et al., 2000; Zwald et al., 2003b; Haskell et al., 2007).
18The reaction norm model was recently introduced to study G × E in animal breeding. This model expresses the phenotype as a polynomial function of the environmental value, where the polynomial coefficients are assumed to be under genetic influence (de Jong, 1995). The reaction norm model is efficient when phenotypes vary continually or gradually over an environmental gradient. It has an analogy with the random regression model, which could explain the recent introduction of the reaction norm model that originates from evolutionary biology in the beginning of the 19th century (Woltereck, 1909), in animal breeding (Strandberg et al., 2000; Calus et al., 2002; Kolmodin et al., 2002). With reaction norm model, covariance functions (Kirkpatrick et al., 1989) are used to model genetic effects over the environmental gradient changes. Breeding values for the coefficients of the function describing the reaction norm and the (co)variances of those coefficients are also estimated. The grouping of environments can be avoided when applying reaction norm model. In addition, the latter model can better explain differences among genotypes in response to external environment parameters (i.e. temperature, humidity). Studies of heat stress and genetic variation in heat tolerance (Ravagnolo et al., 2000; 2002; Bohmanova et al., 2007) are relevant examples.
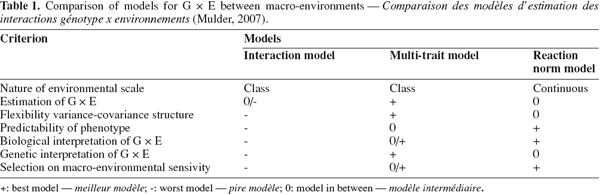
19Mulder (2007) gave a comprehensive review of advantages and disadvantages of models dealing with G x E reported in the literature. The author presented six criteria for using a scoring scale for model comparison (Table 1). The best scored models were the multi-trait and the reaction normal model. de Jong (1995) stated that the reaction norm model is more appropriate for the study of graded responses in continuous environments, whereas the character state model (multi-trait model) is most appropriate to model discrete responses to discrete environments.
4. Genotype by environment in dairy cattle
20Experiments on dairy cattle reported in the literature dealt mainly with genotype by feeding level and system interactions. Two different diets varying in the dry matter proportion of concentrates, brewers’ grains and silage were fed to a herd of two genetically distinct groups based on merit (Veerkamp et al., 1994; Pryce et al., 1999). In general, the G × E interaction exhibited was mostly a scaling effect and there was no significant interaction between diets and lines. In contrast, Veerkamp et al. (1994) reported significantly different regression coefficients of body score condition on pedigree index for fat and protein yield between the two diets. McCarthy et al. (2007a) studied the effect of three Holstein-Friesian strains (high production North American, high durability North American, and New Zealand Holstein-Friesian) and feeding system (high grass allowance feed system, increased stocking rate system, and increased concentrate supplementation) on body weight and body condition score. These authors found that the New Zealand strain remains the most suitable to low cost grass-based system, a predominant system in Ireland. Their study extended previous results found on a subset of the same data and confirmed significant effects of strain of Holstein-Friesian and feed system on reproduction performance (Horan et al., 2004), milk production (Horan et al., 2005), grass dry matter intake (Horan et al., 2006), and somatic cell scores (McCarthy et al., 2007b). All these studies reported important strain by environment interactions. Cows of New Zealand origin produced less milk than North American ones, but had better reproductive performances. Kolver et al. (2002) reported also a re-ranking of New Zealand and North-American genotypes between grazing and mixed ration. Macdonald et al. (2007) compared growth parameters between three different strains of Holstein-Friesian cows grazed on pasture in New Zealand. They concluded that differences in growth parameters and puberty exist among the different genetic strains studied when grazed on pasture.
21In more comprehensive studies (large scale studies), the number of factors differing across environments is large compared to controlled experiments. This is the case in large countries with diverse climatic conditions and production systems. Within country analyses of G × E have been based on modeling data using essentially multi-trait models. Correlations between different environments were used to estimate G × E interactions. König et al. (2005) summarized correlations obtained on intra country analyses for milk traits in temperate countries. Stratification of herds varied by study and was based on: within herd-year mean for mature equivalent milk yield (Castillo-Juárez et al., 2000; Ceron-Munoz et al., 2004), within herd-year standard deviation for mature equivalent milk yield (Raffrenato et al., 2003), regions (Carabaño et al., 1990; Rekaya et al., 2003; König et al., 2005), production level (Calus et al., 2002; Kolmodin et al., 2002), herd size (Gernand et al., 2007), test-day production levels (Veerkamp et al., 1998; Hayes et al., 2003), feeding regimes (Cromie et al., 1998; Boettcher et al., 2003; Fatehi et al., 2003). Estimates of genetic correlations between environments as defined above were high (> 0.80) showing little or no evidence for strong G × E. Almost all the within-country analyses reported only a scaling effect for milk yield with large heterogeneity of variances and in some case heterogeneity of heritability estimates was observed. For example, Boettcher et al. (2003) reported a scaling effect for milk yield with the largest genetic variance and heritability obtained in the conventional systems. The latter authors concluded a lack of presence of G × E between grazing and conventional management systems. In consequence, they reported that selection of sires on grazing systems can be accurate using the national breeding values applied for conventional systems.
22Most of the between-country analyses estimated genetic parameters of traits for each environment studied. However, relationships between traits may also differ by environment. Thus, selection for high production in one environment may lead to different changes in correlated traits under different environments. To avoid this problem, the best way will be to model the relationship between several traits between different environments (Oseni et al., 2004). Recently G × E has been observed for the association of milk yield with protein, fat yield and somatic cell score (Castillo-Juárez et al., 2000; Raffrenato et al., 2003), milk yield with fitness traits (Castillo-Juárez et al., 2000; Windig et al., 2005; Beerda et al., 2007), milk yield with age at first calving (Ruiz-Sánchez et al., 2007). Castillo-Juárez et al. (2000) reported changes in correlations between milk production and somatic cell score and conception rate with specific environments. These correlations were small between pairs of traits in favorable environments and were high in less favorable environments. These authors suggested that an improvement of the management in low environments can reduce the unfavorable correlation found between milk yield and somatic cell score and conception rate.
23The use of germplasm selected in regions with differing climatic conditions and production systems (Bondoc et al., 1989) may result in G × E that could reduce the efficiency of genetic improvement programs in the area where animals will produce. König et al. (2005) summarized genetic correlations for production traits estimated between countries reported in most studies investigating G × E (Table 2). Genetic correlations between the northern hemisphere group (Canada, USA, and Western Europe) ranged between 0.85 and 0.90 (Fikse et al., 2003). Genetic correlations of less than 0.8 were found between North and South America (Stanton et al., 1991; Cienfuegos-Rivas et al., 1999; Costa et al., 2000; Ceron-Munoz et al., 2004) and between some eastern European countries (Rekaya et al., 2001). Low genetic correlations were obtained between countries that differ considerably in climate, management, and production system. Genetic correlation between Mexico and USA was 0.63 (Cienfuegos-Rivas et al., 1999) and 0.49 between Kenya and the United Kingdom (Ojango et al., 2002). Most of these studies pointed out the existence of a scaling effect of G × E, where response to selection was smaller in low input environments than in high input ones. Selection responses to the use of selected US Holstein sires for milk production in Latin America were estimated to range from 53% to 78% of the response observed in the USA (Stanton et al., 1991). In Kenya, the response to selection based on UK breeding values was only 44% (Ojango et al., 2002).
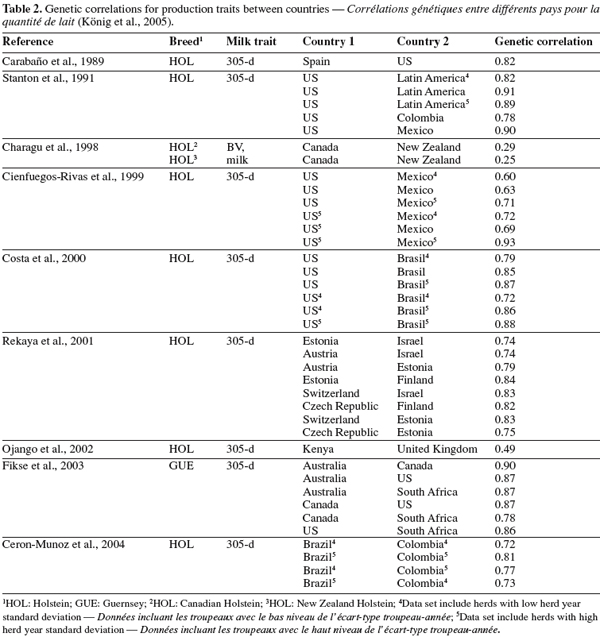
24In a more comprehensive study on between-country analyses, Weigel et al. (2001) found high genetic correlations (> 0.80) between milk yields across 17 Interbull country members. Estimates reported were higher than 0.90 between countries with predominantly grazing systems (i.e. Ireland, Australia, New Zealand). Correlations were also greater than 0.91 between countries with high milk production (US, Canada, Belgium, The Netherlands, and Italy). Correlations between remaining Interbull members ranged between 0.8 and 0.9.
5. Implications of genotype by environment in dairy cattle
25Regardless of the approach applied, the scaling effect of G × E was frequently reported. However, some studies did report the re-ranking effect (Carabaño et al., 1989; Cienfuegos-Rivas et al., 1999; Kolver et al., 2002). In case of scaling effect, animals will maintain their ranking among environments but only differences in the magnitude of breeding values are observed. Pre-adjustment in the data (Wiggans et al., 1991) or correction in the evaluation model (Meuwissen et al., 1996) are able to absorb the scaling effect, and thus the G × E is taken perfectly into account with no consequences on selection decisions. However, weights on traits within a composite index have to be defined with care when a scaling exists for some of these traits (Charagu et al., 1998). In contrast, when re-ranking occurs, superior individuals in one environment will be inferior in other environments. In this case, breeding organizations should face the problem of how to optimize the breeding program to respond to multiple environment requirements. When G × E exists and the environment is under the control of the breeders (i.e. genotype by ration or genetic by housing interaction), it would be easier for breeders to modify the environment to allow optimum expression of the genotype. However, when environments are beyond the breeders’ control, they have to choose the genotypes able to adapt to those environments. One way to accomplish this is the selection of a specific genotype for each environment. This strategy would achieve an optimum response for each environment and help maintain genetic diversity. However, it remains very costly and time consuming to have environment specific genotypes. Furthermore, under these conditions inbreeding may rise and a decline of selection response could be observed. Selection of a trait in one environment with the goal for improving the same trait in other environment known as indirect selection can also be viewed as one of the breeding strategies to address the re-ranking. The efficiency of this selection will depend on the magnitude of the genetic correlation between the two environments and the heritability of the trait in each of the two environments. Togashi et al. (2001a) reported that when sire by country interaction exists, selection of candidate animals in the country with the highest genetic variance should be more effective than selection in the country with the lowest genetic variance. Togashi et al. (2001b) also reported that when G × E interaction is important, an international optimum index becomes more efficient than a within country index as a means to select candidate animals. When considering only sire selection, James (1961) reported that when genetic correlation among two environments was greater than 0.70, testing progeny in both environments and applying a unique index selection were more appropriate than applying separate selection following testing in both environments or selecting and testing in only one of the two environments. Mulder (2007) concluded that a single breeding program with progeny test bulls in both environments was more appropriate when the genetic correlation was higher than 0.60. In contrast, when the genetic correlation was less than or equal to 0.60, it was more opportune to have a specific breeding program and progeny testing in each environment.
6. Conclusion
26This review highlights the importance of G × E in dairy cattle at the animal and breeding programs levels. Estimates of G × E interaction investigated on controlled experiments were low to zero. Advances in statistical modeling of large data sets have allowed good estimates of genetic correlations and heritability of traits in the discrete and the continuous environments. However, difficulties came from the “real” identification of environmental effects. Nevertheless, recent works showed good alternatives based on clustering of environments on “best-definite” descriptors. Practically, all analyses were undertaken in temperate areas with some few investigations between tropical and temperate countries. Evidence on the existence of G × E within or between countries was not clear in some cases. Many studies reported only scaling effects and a few of them reported re-ranking effects. But, nearly all these studies found G × E when differences between environments were large. In diversified intensive production systems, the cost of production, food quality, animal welfare, and consumer desires are all constraints that selection programs should consider. Information on the magnitude of G × E over different time horizons and the “best” environment identification (under favorable and harsh conditions) are needed to help the breeding decision making process. In low input systems, the best alternative to circumvent the consequences of G × E is to select for adaptive traits. This will depend on the genetic correlations between "the import" and "the export" environments.
Bibliographie
Banos G. & Smith C., 1991. Selection within and across populations in livestock improvement. J. Anim. Sci., 69, 2387-2394.
Beerda B. et al., 2007. Effects of genotype by environment interactions on milk yield, energy balance, and protein balance. J. Dairy Sci., 90, 219-228.
Berry D.P. et al., 2007. Genetics of grass dry matter intake, energy balance, and digestibility in grazing Irish dairy cows. J. Dairy Sci., 90, 4835-4845.
Boettcher P.J., Fatehi J. & Schutz M.M., 2003. Genotype x environment interactions in conventional versus pasture-based dairies in Canada. J. Dairy Sci., 86, 383-404.
Bohmanova J., Misztal I. & Cole J.B., 2007. Temperature-humidity indices as indicators of milk production losses due to heat stress. J. Dairy Sci., 90, 1947-1956.
Bondoc O.L., Smith C. & Gibson J.P., 1989. A review of breeding strategies for genetic improvement of dairy cattle in developing countries. Anim. Breed., 57, 819-829.
Calus M.P.L., Groen A.F. & de Jong G., 2002. Genotype x environment interaction for protein yield in Dutch dairy cattle as quantified by different models. J. Dairy Sci., 85, 3115-3123.
Carabaño M.J., Van Vleck L.D., Wiggans G.R. & Alenda R., 1989. Estimation of genetic parameters for milk and fat yields of dairy cattle in Spain and the United States. J. Dairy Sci., 72, 3013-3022.
Carabanõ M.J., Wade K.M. & Van Vleck L.D., 1990. Genotype by environment interactions for milk and fat production across regions of the United States. J. Dairy Sci., 73, 173-180.
Castillo-Juárez H. et al., 2000. Effect of herd environment on the genetic and phenotypic relationships among milk yield, conception rate, and somatic cell score in Holstein cattle. J. Dairy Sci., 83, 807-814.
Ceron-Munoz M.F. et al., 2004. Factors that cause genotype by environment interaction and use of a multiple-trait herd-cluster model for milk yield of Holstein cattle from Brazil and Colombia. J. Dairy Sci., 87, 2687-2692.
Charagu P. & Peterson R., 1998. Estimates of G × E effects for economic efficiency among daughters of Canadian and New Zealand sires in Canadian and New Zealand dairy herds. Interbull Bull., 17, 105-109.
Cienfuegos-Rivas E.G. et al., 1999. Interaction between milk yield of Holstein cows in Mexico and the United States. J. Dairy Sci., 82, 2218-2223.
Cooper M. & DeLacy I.H., 1994. Relationships among analytical methods used to study genotypic variation and genotype-by-environment interaction in plant breeding multi-environment experiments. Theor. Appl. Genet., 88, 561-572.
Costa C.N. et al., 2000. Genetic analysis of Holstein cattle populations in Brazil and the United States. J. Dairy Sci., 83, 2963-2974.
Cromie A.R., Kelleher D.L., Gordon F.J. & Rath M., 1998. Genotype by environment interaction for milk production traits in Holstein Friesian dairy cattle in Ireland. Interbull Bull., 17, 100-104.
de Jong G., 1995. Phenotypic plasticity as a product of selection in a variable environment. Am. Nat., 145, 493-512.
de Jong G. & Bijma P., 2002. Selection and phenotypic plasticity in evolutionary biology and animal breeding. Livest. Prod. Sci., 78, 195-214.
Dickerson G.E., 1962. Implications of genetic environmental interaction in animal breeding. Anim. Prod., 4, 47-64.
Dimov G. et al., 1995. Variance of interaction effects of sires and herd for yield traits of Holsteins in California, New York, and Pennsylvania with an animal model. J. Dairy Sci., 78, 939-946.
Falconer D.S., 1952. The problem of environment and selection. Am. Nat., 86, 293-298.
Falconer D.S. & Mackay T.F.C., 1996. Introduction to quantitative genetics. 4th ed. Harrow, UK: Longman.
Fatehi J., Stella A., Shannon J.J. & Boettcher P.J., 2003. Genetic parameters for feet and leg traits evaluated in different environments. J. Dairy Sci., 86, 661-666.
Fikse W.F., Rekaya R. & Weigel K.A., 2003. Genotype × environment interaction for milk production in Guernsey cattle. J. Dairy Sci., 86, 1821-1827.
Fulkerson W.J., Hough G., Goddard M. & Davison T., 2000. The productivity of Friesian cows: effects of genetic merit and level of concentrate feeding. Final Report – DAN 082. Wollongbar, Australia: Wollongbar Agricultural Institute.
Gernand E., Waßmuth R., von Borstel U.U. & König S., 2007. Heterogeneity of variance components for production traits in large-scale dairy farms. Livest. Prod. Sci., 112, 78-89.
Haskell M.J., Brotherstone S., Lawrence A.B. & White I.M.S., 2007. Characterization of the dairy farm environment in Great Britain and the effect of the farm environment on cow life span. J. Dairy Sci., 90, 5316-5323.
Hayes B.J., Carrick M., Bowman P. & Goddard M.E., 2003. Genotype x environment interaction for milk production of daughters of Australian dairy sires from test-day records. J. Dairy Sci., 86, 3736-3744.
Hill W.G., Edwards M.R., Ahmed M.K.A. & Thompson R., 1983. Heritability of milk yield and composition at different levels and variability of production. Anim. Prod., 36, 59-69.
Horan B. et al., 2004. The effect of strain of Holstein-Friesian cow and feed system on reproductive performance in seasonal-calving milk production systems. Anim. Sci., 79, 453-468.
Horan B. et al., 2005. The interaction of strain of Holstein-Friesian cows and pasture-based feed systems on milk yield, body weight, and body condition score. J. Dairy Sci., 88, 1231-1243.
Horan B. et al., 2006. The effect of strain of Holstein-Friesian dairy cow on grass intake and milk production in various pasture-based systems. Anim. Sci., 82, 435-444.
James J.W., 1961. Selection in two environments. Heredity, 16, 145-152.
Kirkpatrick M. & Heckman N., 1989. A quantitative genetic model for growth, shape, reaction norms, and other infinite-dimensional characters. J. Math. Biol., 27, 429-450.
Kolmodin R.E. et al., 2002. Genotype by environment interaction in Nordic dairy cattle studied using reaction norms. Acta Agric. Scand. A Anim. Sci., 52, 11-24.
Kolver E.S. et al., 2002. Total mixed rations versus pasture diets: evidence for a genotype x diet interaction in dairy cow performance. Proc. N.Z. Soc. Anim. Prod., 62, 246-251.
König S., Dietl G., Raeder I. & Swalve H.H., 2005. Genetic relationships for dairy performance between large-scale and small-scale farm conditions. J. Dairy Sci., 88, 4087-4096.
Lin C.Y. & Togashi K., 2002. Genetic improvement in the presence of genotype by environment interaction. Anim. Sci. J., 73, 3-11.
Lohuis M.M. & Dekkers J.C.M., 1998. Merits of borderless evaluations. In: Proceeding of the 6th world congress on genetics applied to livestock production, January 11-16, 1998, Armidale, Australia, 169-172.
Macdonald K.A. et al., 2007. A comparison of three strains of Holstein-Friesian cows grazed on pasture: growth, development, and puberty. J. Dairy Sci., 90, 3993-4003.
McCarthy S. et al., 2007a. Influence of Holstein-Friesian strain and feed system on body weight and body condition score lactation profiles. J. Dairy Sci., 90, 1859-1869.
McCarthy S. et al., 2007b. Effect of strain of Holstein-Friesian and feed system on udder health and milking characteristics. Livest. Prod. Sci., 107, 19-28.
Meuwissen T.H.E., de Jong G. & Engel B., 1996. Joint estimation of breeding values and heterogeneous variances of large data files. J. Dairy Sci., 79, 310-316.
Mulder H.A., 2007. Methods to optimize livestock breeding programs with genotype by environment interaction and genetic heterogeneity of environmental variance. PhD thesis: Wageningen University (The Netherlands).
Nauta W.J., Veerkamp R.F., Brascamp E.W. & Bovenhuis H., 2006. Genotype by environment interaction for milk production traits between organic and conventional dairy cattle production in The Netherlands. J. Dairy Sci., 89, 2729-2737.
Ojango J.M.K. & Pollot G.E., 2002. The relationship between Holstein bull breeding values for milk yield derived in both the UK and Kenya. Livest. Prod. Sci., 74, 1-12.
Oseni S., Misztal I., Tsuruta S. & Rekaya R., 2004. Genetic components of days open under heat stress. J. Dairy Sci., 87, 3022-3028.
Ouweltjes W. et al., 2007. Effects of management and genetics on udder health and milk composition in dairy cows. J. Dairy Sci., 90, 229-238.
Powell R.L. & Sieber M., 1994. The origin of the world's best sires. Holstein Friesian J., June, 427-440.
Pryce J.E., Nielson B.L., Veerkamp R.F. & Simm G., 1999. Genotype and feeding system effects and interactions for health and fertility traits in dairy cattle. Livest. Prod. Sci., 57, 193-201.
Raffrenato E. et al., 2003. Genotype by environment interaction for yield and somatic cell score with alternative environmental definitions. J. Dairy Sci., 86, 2470-2479.
Ravagnolo O., Misztal I. & Hoogenboom G., 2000. Genetic component of heat stress in dairy cattle, development of heat index function. J. Dairy Sci., 83, 2120-2125.
Ravagnolo O. & Misztal I., 2002. Studies on genetics of heat tolerance in dairy cattle with reduced weather information via cluster analysis. J. Dairy Sci., 85, 1586-1589.
Rekaya R., Weigel K.A. & Gianola D., 2001. Application of a structural model for genetic covariances in international dairy sire evaluations. J. Dairy Sci., 84, 1525-1530.
Rekaya R., Weigel K.A. & Gianola D., 2003. Bayesian estimation of parameters of a structural model for genetic covariances between milk yields in five regions of the United States. J. Dairy Sci., 86, 1837-1844.
Ruiz-Sánchez R. et al., 2007. Changes in the association between milk yield and age at first calving in Holstein cows with herd environment level for milk yield. J. Dairy Sci., 90, 4830-4834.
Schaeffer L.R., 1994. Multiple-country comparison of dairy sires. J. Dairy Sci., 77, 2671-2678.
Stanton T.L., Blake R.W. & Quaas R.L., 1991. Genotype by environment interaction for Holstein milk yield in Colombia, Mexico and Puerto Rico. J. Dairy Sci., 74, 1700-1714.
Strandberg E. et al., 2000. Genotype by environment interaction in Nordic dairy cattle studied by use of reaction norms. Interbull Bull., 25, 41-45.
Togashi K. et al., 2001a. Partition of sire effects for international sire evaluation in the presence of genotype-environment interaction. Livest. Prod. Sci., 73, 225-235.
Togashi K., Lin C.Y. & Moribe K., 2001b. Construction of optimum index to maximize overall performance across countries in the presence of genotype-environment interaction. J. Dairy Sci., 84, 1872-1883.
Tollens E., Matty D. & Swennen R., 2004. Agrobiotechnology in developing countries: North-South partnerships are the key. Outlook Agric., 33, 231-238.
Van der Werf J.H.J. & Ten Napel J., 1991. Estimation of genotype by environment interaction for milk production under Dutch circumstances. In: Proceeding of the 42nd annual meeting of EAAP, September 8-12, 1991, Berlin, Germany, 213-218
Veerkamp R.F., Simm G. & Oldham J.D., 1994. Effects of interaction between genotype and feeding system on milk production, feed intake, efficiency and body tissue mobilization in dairy cows. Livest. Prod. Sci., 39, 229-241.
Veerkamp R.F., Simm G. & Oldham J.D., 1995. Genotype by environment interaction: experience from Langhill. In: Lawrence T.L., Gordon F.J. & Carson A., eds. Breeding and feeding the high genetic merit dairy cow. Midlothian, Scotland: British Society of Animal Science Occasional Publications, 59-66.
Veerkamp R.F. & Goddard M.E., 1998. Covariance functions across herd production levels for test day records on milk, fat, and protein yields. J. Dairy Sci., 81, 1690-1701.
Via S. et al., 1995. Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol. Evol., 10, 212-217.
Weigel K.A. & Rekaya R., 2000. A multiple-trait herd cluster model for international dairy sire evaluation. J. Dairy Sci., 83, 815-821.
Weigel K.A., Rekaya R., Zwald N.R. & Fikse W.F., 2001. International genetic evaluation of dairy sires using a multiple-trait model with individual animal performance records. J. Dairy Sci., 84, 2789-2795.
Wiggans G.R. & Van Raden P.M., 1991. Method and effect of adjustment for heterogeneous variance. J. Dairy Sci., 74, 4350-4357.
Windig J.J., Calus M.P.L. & Veerkamp M.F., 2005. Influence of herd environment on health and fertility and their relationship with milk production. J. Dairy Sci., 88, 335-347.
Woltereck R., 1909. Weiterer experimentelle Untersuchunger uber Artveranderung, Speziell uber das Wessen Quantitativer Artunterschiede bei Daphniden. Dtsch Zool. Ges., 19, 110-172.
Zwald N.R., Weigel K.A., Fikse W.F. & Rekaya R., 2003a. Identification of factors that cause genotype by environment interaction between herds of Holstein cattle in seventeen countries. J. Dairy Sci., 86, 1009-1018.
Zwald N.R., Weigel K.A., Fikse W.F. & Rekaya R., 2003b. Application of a multiple-trait herd cluster model for genetic evaluation of dairy sires from seventeen countries. J. Dairy Sci., 86, 376-382.
Om dit artikel te citeren:
Over : Hedi Hammami
Gembloux Agricultural University – FUSAGx. Animal Science Unit. Passage des Déportés, 2. B-5030 Gembloux (Belgium). E-mail: hammami.h@fsagx.ac.be – Office de l’Elevage et des Paturages. Rue Alain Savary, 30. TN-1002 Tunis-Belvedere (Tunisia).
Over : Boulbaba Rekik
Ecole Supérieure d’Agriculture de Mateur. TN-7030 Mateur (Tunisia).
Over : Nicolas Gengler
Gembloux Agricultural University – FUSAGx. Animal Science Unit. Passage des Déportés, 2. B-5030 Gembloux (Belgium). – National Fund for Scientific Research. Rue d’Egmont, 5. B-1000 Brussels (Belgium).






