- Accueil
- volume 13 (2009)
- numéro 4
- Phylogenetic diversity of Moroccan cork oak woodlands fungi
Visualisation(s): 0 (0 ULiège)
Téléchargement(s): 0 (0 ULiège)
Phylogenetic diversity of Moroccan cork oak woodlands fungi

Notes de la rédaction
Received on January 28, 2009; accepted on May 8, 2009
Résumé
Diversité phylogénétique des champignons des subéraies marocaines. La variabilité interspécifique de 87 carpophores des champignons appartenant à 15 genres et 39 espèces ont été évaluées par l'analyse du polymorphisme de longueur de fragments de restriction (RFLP) de l'espaceur interne transcrit (ITS). Cette région a été d'abord amplifiée, avec des amorces spécifiques, par la réaction de polymérisation en chaine (PCR), puis digérée avec différentes enzymes de restriction. Un produit d'amplification, dont la taille variait entre 500 et 950 paires de bases (pb), a été obtenu pour tous les isolats analysés. Le degré de polymorphisme observé ne permet pas l'identification de la plupart des espèces. L’analyse RFLP, avec les enzymes de restriction Alu I, EcoR I et Hinf I, des fragments amplifiés révèle un grand polymorphisme. Les quinze genres et la plupart des espèces présentent des profils de restriction spécifiques. Les deux espèces du genre Russula (R. decipiens et R. straminea) présentent le même profil de restriction. Ces espèces pourraient être considérées comme étroitement apparentées. Les carpophores de Pisolithus montrent deux profils ITS-RFLP distincts. Le séquençage de l’ITS confirme que ces deux profils correspondent à deux espèces distinctes de Pisolithus. Nos données montrent l’importance de l’analyse PCR-RFLP de l’ITS pour la caractérisation moléculaire et l'identification des champignons ectomycorhiziens et leur suivi dans les programmes d'inoculation artificielle.
Abstract
Interspecific variation among 87 sporocarps of fungi belonging to 15 genera and 39 species were evaluated by analyzing the internal transcribed spacer (ITS) of the rDNA region using restriction fragment length polymorphism (RFLP). The ITS region was first amplified by polymerase chain reaction (PCR) with specific primers and then cleaved with different restriction enzymes. Amplification products, which ranged between 500 and 950 base pairs (bp), were obtained for all the isolates analyzed. The degree of polymorphism observed did not allow proper identification of most of the species. Cleavage of amplified fragments with the restriction enzymes Alu I, EcoR I and Hinf I revealed extensive polymorphism. The fifteen genera and most species presented specific restriction patterns. The only species not identifiable by a specific pattern belonged to the genera Russula (R. decipiens and R. straminea). These species might be considered as closely related species. The Pisolithus sporocarps had two ITS-RFLP types with one dominating. ITS sequencing confirms that the two RFLP types correspond to two distinct species of Pisolithus. Our data show the potential of ITS region PCR-RFLP for the molecular characterization of ectomycorrhizal fungi and their identification and monitoring in artificial inoculation programs.
Table des matières
1. Introduction
1In Morocco, the cork oak woodlands of Maâmora forest undergo a worrying regression in spite of the intensive plantation programs. The success of these programs with a good plant establishment after transfer to the field, is related among others to the cultural techniques used in nurseries. Ectomycorrhizal symbiosis, a mutualistic plant-fungus association, plays a fundamental role in the biology and ecology of forest trees, affecting growth, water and nutrient absorption, and providing protection from root diseases (Smith et al., 1997). The controlled mycorrhization in nurseries, by selected ectomycorrhizal fungi, improves survival, establishment, and growth of seedlings after outplanting (e.g. Marx, 1977; Le Tacon et al., 1992). It is estimated that between 5,000 and 6,000 species of fungi worldwide form ectomycorrhizas (Molina et al., 1992). Most of these are basidiomycetes, but a small number of ascomycetes and zygomycetes are also known to produce this type of symbiosis (Smith et al., 1997). Phenotypic differences between two isolates of the same species of an ectomycorrhizal fungus may be as pronounced as the differences between two distinct species, and for this reason identification of these fungi is not clear-cut (De la Bastide et al., 1995). Aboveground surveys of sporocarps are usually poor indicators of the community structure belowground (Arnolds, 1991; Jonsson et al., 2000; Horton et al., 2001). This is because sporocarp production is triggered by specific environmental conditions (Eggers, 1995; Gardes et al., 1991a). A classical approach for identifying mycorrhizas is therefore to trace hyphal connections between sporocarps and mycorrhizal sheaths (Agerer, 1995). However, special skills are required when root density is so high that hyphae emerging from the stipe base cannot be attributed unambiguously to a single mycorrhizal type (Pritsch, 1996). An alternative promising way to identify mycorrhizas consists of comparing specific DNA regions of mycorrhizas and sporocarps. Polymerase chain reaction coupled with restriction fragment length polymorphism analyses (PCR/RFLP) have been applied in mycorrhizal research to identify strains of introduced or naturally occurring mycorrhizal fungi (Gardes et al., 1991a; Henrion et al., 1992; Gardes et al., 1993; Henrion et al., 1994a), or of economically important species such as Tuber (Henrion et al., 1994b), and also to differentiate and identify mycorrhizal symbionts unambiguously (Erland et al., 1994; Kraigher et al., 1995; Gardes et al., 1996a). In these studies, two target sequences within the ribosomal DNA, ITS (Internal Transcribed Spacer) and IGS (Intergenic Spacer), have mainly been used to detect polymorphism between and within species of ectomycorrhizal taxa. The ITS region separating genes 17S and 25S can be amplified by specific primers anchored in these two units. Since the ITS region is highly conserved intraspecifically but variable between different species it is often used in taxonomy (Bruns et al., 1991; Hillis et al., 1991), but ITS region polymorphism of identifying ectomycorrhizal fungi species has been determined for only a restricted number of species, leaving its full potential as a taxonomic tool as yet unexplored (Karén et al., 1997). The PCR/RFLP polymorphism of the ITS region is generally regarded as appropriate to differentiate at the species level (Eggers, 1995; Gardes et al., 1996b). Martin et al. (1998), based on PCR/RFLP of the rDNA ITS, have been set up an expandable database of DNA profiles and fingerprints of ectomycorrhizal fungi in a World Wide Web (WWW) Internet Server.
2In this study, 87 sporocarps of known fungi belonging to 15 genera and 39 different species were analyzed using the PCR/RFLP technique as applied to the ITS regions of these fungi. These analyses aimed to confirm the classification of these fungi and to elucidate their interspecies biodiversity. The ultimate aim of our investigation is to establish a database of DNA profiles of Moroccan cork oak woodlands ectomycorrhizal fungi to help identification of different morphotypes found in the field.
2. Materials and methods
2.1. Sporocarp collections
3Sporocarps were collected from eight different Moroccan cork oak woodlands all located at the northern part of Morocco. Data are presented only for well identified ones according to distinct macroscopic and microscopic characters (Table 1).

2.2. Isolates cultivation
4Pure cultures were obtained by transferring aseptically pieces of fruit bodies on the Modified Melin Norkrans (MNM) agar medium (Marx et al., 1975). They were incubated on the culture medium in the dark for 25 days at 28°C. All the isolates are conserved within the INRA (Montpellier, France) and the Forest Research Centre (Rabat, Morocco) ectomycorrhizal fungi culture collections, on MNM under 2°C.
2.3. Total DNA extraction and PCR amplification of ITS rDNA
5Total DNA was extracted from fruiting bodies using the CTAB protocol (Gardes et al., 1993; Henrion et al., 1994b) or the DNeasy Plant Mini Kit according to the manufacturer’s recommendations (QIAgen SA). The internal transcribed spacer (ITS) of the nuclear ribosomal DNA (rDNA) was amplified by using ITS1 and ITS4 primers (White et al., 1990). PCR amplification was carried out using a PCR-100 thermocycler (MJ Research, Inc. Watertown, MA, USA) using 50 µL reaction volumes each containing: 1 µL DNA template, 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 200 µM of each dNTP, 0.5 µM of each primer and 0.6 units of Taq DNA polymerase (Eurogentec, Belgium). The amplification conditions were 1 cycle of 95°C for 5 min, followed by 35 cycles including 94°C for 30 s, 55°C for 30 s, 72°C for 1 min with a final extension at 72°C for 7 min. Negative controls (no DNA template) were included in all PCR experiments to check for DNA contamination.
2.4. RFLP analysis of ITS diversity
6Per each 10 µL restriction digest, 8 µL amplified ITS product was mixed with the appropriate restriction buffer and 5 units of the appropriate endonucleases Alu I, EcoR I and Hinf I (Gibco BRL, Life Technologies) and then incubated for at least 2 h at 37°C. ITS and digested ITS products were respectively migrated by electrophoresis on 2% and on 3% regular (Sigma) and Nusieve (FMC) agarose gels, stained with ethidium bromide and photographed using the Oncor-Appligene Imager 2.02. Digested 100 bp (molecular weight marker, Boehringer Mannhein) and Smartladder (Eurogentec, Belgium) were used as ladder.
2.5. Data analysis
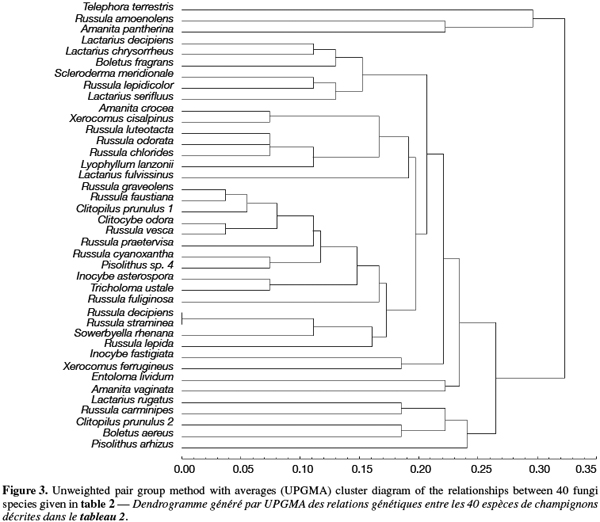
7After restriction with endonucleases, the amplification products were scored as 1 (presence) or 0 (absence) of a restriction site and used for determining genetic distances between the isolates (Nei et al., 1979). These distances were used to cluster the isolates by the unweighted pair group method with averages (UPGMA) method using the Statistica program (version 4.5 for Windows, StatSoft, Inc. 1993, Tulsa, OK, USA).
3. Results
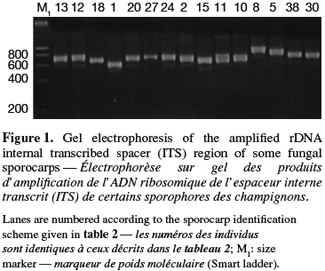
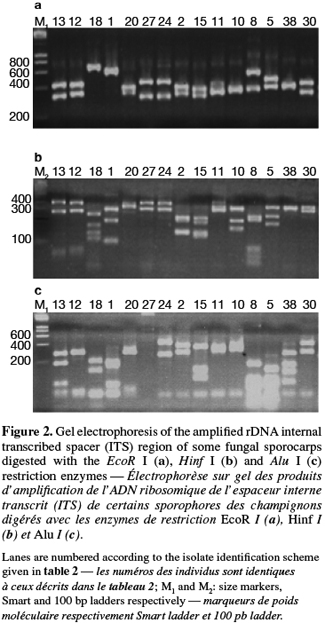
8A total of 87 sporocarps in 15 genera and 39 species were analyzed, with a range of 1-3 sporocarps of all the species except for Pisolithus spp. and Boletus aereus represented by 19 and 16 sporocarps, respectively. PCR amplification with specific primers for the ITS region generated bands ranging from 500 to 950 bp. Figure 1 shows that all samples presented only one amplification product. In general, no difference was detected between species from the same genus. To detect a wider range of polymorphism, the PCR products were cleaved with the restriction enzymes Alu I, EcoR I and Hinf I. The Alu I restriction enzyme produced 39 ITS-RFLP patterns including those of Pisolithus spp., while EcoR I and Hinf I generated 35 and 34 patterns, respectively. The restriction fragment sizes of ITS digested with these respective enzymes are summarized in table 2 and examples of ITS-RFLP profiles are depicted in figure 2a, 2b and 2c. All the species showed polymorphic patterns, except two species of Russula (R. decipiens and R. straminea) which showed the same RFLP type for the three endonucleases. For this reason, the PCR products from these Russula species were further cleaved with two other enzymes (Hae III, Taq I) but still no polymorphism was detected. Furthermore, all the sporocarps of the same species (e.g. Boletus aereus, Russula amoenolens, R. decipiens and R. straminea) collected at local and large scale, except Pisolithus spp. and Clitopilus prunulus, yielded a single RFLP type. By using the enzymes Alu I, EcoR I and Hinf I, two patterns have been observed within Pisolithus and Clitopilus prunulus sporocarps; only one of these restriction enzymes is enough to elucidate this polymorphism. A comparison of Pisolithus ITS sequences using the BLAST program showed that the ITS-RFLP type 1 and type 2 were respectively very similar (96-98% of sequence similarities) to ITS of species 6 and species 4 defined by Martin et al. (2002). The ITS-RFLP type 1 was the dominant pattern representing 97% of collected sporocarps. The neighbor-joining tree separated the isolates into two groups (data not shown). Group I contained ITS-RFLP type 1 from Pisolithus arrhizus named species 6, while group II contained ITS-RFLP type 2 from Pisolithus sp. named species 4 (Martin et al., 2002). Restriction fragments obtained with all the endonucleases tested were used to determine genetic distances between the genotypes and cluster them into specific groups. The phylogenetic tree constructed is depicted in figure 3. In general, no genera specific group was detected. Except for R. decipiens and R. straminea which considered as closely related species and some species of the same genera which formed specific group, most of the species were clustered independently of their belonging to a genus.
4. Discussion
9Mycorrhizal inoculation of seedlings with selected ectomycorhizal fungi is now applied worldwide to improve the survival, establishment and growth of seedlings after outplanting (Le Tacon et al., 1992; Grove et al., 1993). Morphological descriptions (Agerer, 1991) or allozymes (Sen, 1990) have provided useful data for identifying the mycorrhizal fungi below ground, but most species have not been described by these methods. Today a wide range of molecular techniques can be used to detect DNA sequence variation in ectomycorrhizal fungi (Gardes et al., 1991a; 1991b; Henrion et al., 1992; Bruns et al., 1993).
10Detection of polymorphism using PCR-RFLP analyses of the rDNA ITS region has been successfully used for identifying several species of fungi (Gardes et al., 1996a; Amicucci et al., 1996; Di Battista, 1997; Karen et al., 1997; Pritsch et al., 1997). This simple technique requires only minute amounts of DNA and two specific primers flanking the ITS region. We found that the amplification products for the ITS region of 39 species (100%) of fungi collected in Moroccan cork oak woodlands ranging from 500 to 950 bp, coincided with the sizes obtained for the other fungi (Gardes et al., 1991a; Karen et al., 1997; Martin et al., 1998). Despite the length polymorphism observed for many of the species, ITS analysis alone was not able to separate all the genotypes. RFLP analysis of the ITS region has been suggested by several authors as a means for discriminating between fungi at the interspecific and intraspecific level (Bruns et al., 1991; Gardes et al., 1990; 1991b; Guerin-Laguette, 1998; Manassila et al., 2005). Cleavage of the ITS region with Alu I (Figure 2) allowed differentiation of 40 out of the 39 identified species according to macroscopic and microscopic characteristics (Table 2). In fact, two ITS-RFLP types of Pisolithus are identified and ITS sequence analysis indicated that the two ITS-RFLP types correspond to two distinct Pisolithus species, species 6 and species 4, recognized by Martin et al. (2002). Furthermore, none of the restriction enzymes produced a distinct pattern for the 2 Russula species (R. decipiens and R. straminea). Henrion et al. (1992) considered those types of species, which do not differ enough from the ITS region to be distinguished with this marker (e.g. Laccaria bicolor and Laccaria laccata) as closely related species.
11A correlation between soil properties and diversity within the genus Pisolithus have been revealed (Bakkali Yakhlef et al., 2007; 2008). Due to this high degree of interspecific variation of the ITS, a matching of RFLP types of mycorrhizas and identified sporocarps (reference) RFLP types within the same geographical area will be likely to indicate identical species. Basing on PCR/RFLP of ITS of ectomycorrhizal fungi, a RFLP database has been setting up in a World Wide Web Internet server (Martin et al. 1998).
12Community studies of ectomycorrhizal fungi are based either on identification of mycorrhizas (the so-called below-ground view), or on monitoring of fruit body production (above-ground view) (Richard et al., 2005). It is clear that fruit body surveys reveal the presence of ECM taxa in a fast and inexpensive way (Richard et al., 2004). However these studies often underestimate the presence of numerous taxa, like resupinate and hypogeous fungi and taxa lacking an apparent sexual stage (Horton et al., 2001). Cenococcum geophilum, for example, which is an abundant, cosmopolitan species lacking sexual structures and reproduces via sclerotia and/or mycelia (Jonsson et al., 2000; Horton et al., 2001), has been found in abundance, blow ground, in cork oak woodlands (Abourouh, 1991).
13The results outlined in this paper show that interspecific variation of the ectomycorrhizal fungi ITS region is relatively high. ITS restriction fragment analysis has potential for developing species-level markers for many, but not necessarily all, ectomycorrhizal fungi species. It appears that ITS-RFLP is a potent tool for the taxonomic study of ectomycorrhizal fungi, with the minute amounts of DNA required and the high reproducibility of this procedure making it an ideal method both for studying population heterogeneity in the field and the identification and monitoring of specific strains introduced into the soil in controlled mycorrhization programs.
14Acknowledgements
15This work was supported by the PRAD (Projet de Recherche Agronomique pour le Développement) 04-13 " Diversité, écologie et utilisation des Pisolithus spp. pour la gestion durable des subéraies marocaines (Pisum) ". The authors thank Dr. Pierre-Arthur Morreau for helping them in the identification of fungi in the field.
Bibliographie
Abourouh M., 1991. La distribution au Maroc de Cenococcum geophilum Fr. Ann. Rech. For. Maroc, 25, 30-40.
Agerer R., 1991. Characterization of ectomycorrhiza. In: Norris J.R., Read D.J. & Varma A.K., eds. Methods in microbiology. London: Academic Press, 25-73.
Agerer R., 1995. Anatomical characteristics of identified ectomycorrhizas: an attempt towards a natural classification. In: Varma A. & Hock B., eds. Mycorrhiza. Berlin; Heidelberg, Deutschland; New York, USA: Springer.
Amicucci A. et al., 1996. Identification of ectomycorrhizae from Tuber species by RFLP analysis of the ITS region. Biotechnol. Lett., 18, 821-826.
Arnolds E., 1991. Decline of ectomycorrhizal fungi in Europe. Agric. Ecosystems Environ., 35, 209-244.
Bakkali Yakhlef S. et al., 2007. Analyse de la variabilité intraspécifique des Pisolithus spp. du Chêne-liège au Maroc. In : Communication 2e Assises de la recherche forestière, Kénitra, Maroc.
Bakkali Yakhlef S. et al., 2008. Analyse moléculaire, par PCR-RFLP, de la diversité génétique des champignons ectomycorhiziens des subéraies marocaines : cas de Pisolithus spp. Ann. Rech. For. Maroc, 39, 58-62.
Bruns T.D., White T.J. & Taylor J.W., 1991. Fungal molecular systematics. Ann. Rev. Ecol. Syst., 22, 525-564.
Bruns T. & Gardes M., 1993. Molecular tools for the identification of ectomycorrhizal fungi-taxon-specific oligonucleotide probes for suilloid fungi. Mol. Ecol., 2, 233-242.
De la Bastide P.Y., Kropp B.R. & Piché Y., 1995. Population structure and mycelial phenotypic variability of the ectomycorrhizal basidiomycete Laccaria bicolor (Maire) Orton. Mycorrhiza, 5, 371-379.
Di Battista C., 1997. Comportement en pépinière et en plantation d’un champignon ectomycorhizien Laccaria bicolor S238N inoculé sur Epicéa (Picea abies) et Douglas (Pseudotsuga menziesii). Typage moléculaire de la souche introduite. Thèse de doctorat : Université Henri Poincarré de Nancy (France).
Eggers K.N., 1995. Molecular analysis of ectomycorrhizal communities. Can. J. Bot., 73, 1415-1422.
Erland S. et al., 1994. Identification of the ectomycorrhizal basidiomycete Tylospora fibrillosa Donk by RFLP analysis of the PCR-amplified ITS and IGS regions of ribosomal DNA. New Phytol., 126, 525-532.
Gardes M. & Bruns T., 1993. ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Mol. Ecol., 2, 113-118.
Gardes M. & Bruns T.D., 1996a. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: above- and below-ground views. Can. J. Bot., 74, 1572-1583.
Gardes M. & Bruns T.D., 1996b. ITS-RFLP matching for the identification of fungi. In: Clapp J.P., ed. Methods in molecular biology: species diagnostic protocols: PCR and other nucleic acid methods. Totowa, NJ, USA: Humana Press Inc., 177-186.
Gardes M., Fortin J.A., Mueller G.M. & Kropp B.R., 1990. Restriction fragment length polymorphisms in the nuclear ribosomal DANN of four Laccaria spp.: L. bicolour, L. laccata, L. proxima, and L. amethystina. Phytopathology, 80, 1312-1317.
Gardes M., Mueller G.M., Fortin J.A. & Kropp B.R., 1991a. Mitochondrial DNA polymorphisms in Laccaria bicolor, L. Laccat, L. Proxima and L. amethystina. Mycolog. Res., 95, 206-216.
Gardes M. et al., 1991b. Identification of indigenous and introduced symbiotic fungi in ectomycorrhizae by amplification of nuclear and mitochondrial ribosomal DNA. Can. J. Bot., 69, 180-190.
Grove T.S. & Le Tacon F., 1993. Mycorrhiza in plantation forestry. Adv. Plant Pathol., 23, 191-227.
Guerin-Laguette L., 1998. Les lactaires à lait rouge : mycorrhization contrôlée des pins et caractérisation moléculaire. Application à l’étude de la compétence écologique et de la compétitivité d’isolats de Lactarius delicious. Thèse de doctorat : Ecole Nationale Supérieure Agronomique de Montpellier (France).
Henrion B., Chevalier G. & Martin F., 1994a. Typing truflle species by PCR amplification of the ribosomal DNA spacers. Mycol. Res., 98, 37-43.
Henrion B., Di Battista C. & Bouchard D., 1994b. Monitoring the persistence of Laccaria bicolor as an ectomycorrhizal symbiont of nursery-grown Douglas fir by PCR of the rDNA intergenic spacer. Mol. Ecol., 3, 571-580.
Henrion B., Le Tacon F. & Martin F., 1992. Rapid identification of genetic variation of ectomycorrhizal fungi by amplification of ribosomal RNA genes. New Phytol., 122, 289-298.
Hillis D.M. & Dixon M.T., 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol., 66, 411-453.
Horton T.R. & Bruns T.D., 2001. The molecular revolution in ectomycorrhizal ecology: peeking into the black-box. Mol. Ecol., 10, 1855-1871.
Jonsson L., Dahlberg A. & Brandrud T.E., 2000. Spatio-temporal distribution of an ectomycorrhizal community in an oligotrophic Swedish Picea abies forest subjected to experimental nitrogen addition: Above- and below-ground views. Forest Ecol. Manage., 132, 143-156.
Karén O. et al., 1997. Inter- and intraspecific variation in the ITS region of rDNA of ectomycorrhizal fungi in Fennoscandia as detected by endonuclease analysis. New Phytol., 136, 313-325.
Kraigher H., Agerer R. & Javornik B., 1995. Ectomycorrhizae of Lactarius lignyotus on Norway spruce, characterized by anatomical and molecular tools. Mycorrhiza, 5, 175-180.
Le Tacon F. et al., 1992. Variations in field response of forest trees to nursery ectomycorrhizal inoculation in Europe. In: Read D.J., Lewis D., Fitter A. & Alexander I., eds. Mycorrhizas in ecosystems. Wallington, UK: CAB International, 119-134.
Manassila M. et al., 2005. Phylogenetic diversity of wild edible Russula from Northeastern Thailand on the basis of internal transcribed spacer sequence. Sci. Asia, 31, 323-328.
Martin F. et al., 1998. Molecular markers in ecology of ectomycorrhizal fungi. Genet. Sel. Evol., 30(Suppl. 1), 333-355.
Martin F., Diez J., Dell B. & Delaruelle C., 2002. Phylogeography of the ectomycorrhizal Pisolithus species as infered from nuclear ribosomal DNA ITS sequences. New Phytol., 153, 345-357.
Marx D.H., 1977. Tree host range and world distribution of the ectomycorrhizal fungus Pisolithus tinctorius. Can. J. Microbiol., 23, 217-233.
Marx D.H. & Bryan W.C., 1975. Growth and ectomycorrhizal development of loblolly pine seedlings in fumigated soil infested with the fungal symbiont Pisolithus tinctorius. Forest Sci., 21, 245-254.
Molina R.H., Massicotte H. & Trappe J., 1992. Specificity phenomena in mycorrhizal symbiosis: community-ecological consequences and practical implications. In: Allen M.F., ed. Mycorrhizal functioning. London, UK: Springer.
Nei M. & Li W.H., 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA, 76, 5269-5273.
Pritsch K., 1996. Studies on diversity and ecology of the black mycorrhiza (Alnus glutinosa (L.) Gaertn.). PhD Thesis: University of Tuebingen (Germany).
Pritsch K., Boyle H., Munch J.C. & Buscot F., 1997. Characterization and identification of black alder ectomycorrhizas by PCR/RFLP analyses of the rDNA internal transcribed spacer (ITS). New Phytol., 137, 357-369.
Richard F., Moreau P.A., Selosse M.A. & Gardes M., 2004. Diversity and fruiting patterns of ectomycorrhizal and litter saprobic fungi in an old-growth Mediterranean forest dominated by Quercus ilex L. Can. J. Bot., 82, 1711-1729.
Richard F., Millot S., Gardes M. & Selosse M.A., 2005. Diversity and specificity of ectomycorrhizal fungi retrieved from an old-growth Mediterranean forest dominated by Quercus ilex. New Phytol., 166, 1011-1023.
Sen R., 1990. Intraspecific variation in two species of Suillus from scots pine (Pinus sylvestris L.) forests based on somatic incompatibility and isozyme analyses. New Phytol., 114, 607-616.
Smith S.E. & Read D.J., 1997. Mycorrhizal symbiosis. 2nd ed. London: Academic Press.
White T.J., Bruns T.D., Lee S.S. & Taylor J.W., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelf D.H., Sninsky J.J. & White T.J., eds. PCR protocols: a guide to methods and application. San Diego, USA: Academic Press, 315-322.
Pour citer cet article
A propos de : Salaheddine Bakkali Yakhlef
Centre de Recherche Forestière. Charia Omar Ibn Khattab. B.P. 763 Agdal. MA-Rabat (Maroc). E-mail: bakkali_yse@yahoo.fr
A propos de : Benaissa Kerdouh
Centre de Recherche Forestière. Charia Omar Ibn Khattab. B.P. 763 Agdal. MA-Rabat (Maroc).
A propos de : Daniel Mousain
Institut National de la Recherche Agronomique (INRA). Unité sous contrat (USC) Symbioses Tropicales et Méditerranéennes. F-34398 Montpellier (France).
A propos de : Marc Ducousso
Institut Agronomique Néo-Calédonien (IAC). BPA5. F-98848 Nouméa (Nouvelle-Calédonie).
A propos de : Robin Duponnois
Institut de Recherche pour le Développement (IRD). Laboratoire commun de Microbiologie IRD/ISRA/UCAD. Centre de Recherche de Bel Air. BP 1386. SN-Dakar (Sénégal).
A propos de : Mohamed Abourouh
Centre de Recherche Forestière. Charia Omar Ibn Khattab. B.P. 763 Agdal. MA-Rabat (Maroc).






