- Portada
- Volume 17 (2013)
- numéro 1
- Ribosome Inactivating Protein of barley enhanced resistance to Rhizoctonia solani in transgenic potato cultivar ‘Desirée’ in greenhouse conditions
Vista(s): 0 (0 ULiège)
Descargar(s): 0 (0 ULiège)
Ribosome Inactivating Protein of barley enhanced resistance to Rhizoctonia solani in transgenic potato cultivar ‘Desirée’ in greenhouse conditions

Notes de la rédaction
Received on September 14, 2011; accepted on November 8, 2012
Résumé
Rip30 améliore la résistance au Rhizoctonia solani chez les plantes transgéniques de pomme de terre cultivar ‘Désirée’ en conditions de serre. Dans la présente étude, le cultivar ‘Désirée’ de pomme de terre (Solanum tuberosum L.) a été transformé en utilisant la souche LBA4404 d'Agrobacterium tumefaciens renfermant le plasmide pBIN19 où est cloné le gène rip30 (Ribosome Inactivating Protein). Des disques de feuilles ont été utilisés comme explants pour la transformation. Les paramètres de régénération in vitro qui ont été évalués sont le pourcentage de cals régénérés, le nombre de pousses par cal, le pourcentage d’enracinement des pousses et le pourcentage de plantes transgéniques. La technique PCR a été utilisée pour l'identification des plantes transformées. Les analyses par Southern blot et Western blot ont été utilisées pour la caractérisation moléculaire des transformants. Une inoculation dans des conditions de serre a été réalisée pour évaluer la résistance au pathogène Rhizoctonia solani des clones transgéniques exprimant le gène rip30. Les résultats de cette étude ont révélé que les plantes développées en milieu sélectif n’ont pas toutes été positives pour le gène correspondant en utilisant la technique de PCR. L’analyse Southern blot a confirmé que les plantes transgéniques ont intégré trois copies du gène rip30 dans leur génome. L'expression de la protéine RIP30 a été confirmée pour les clones transgéniques par l’analyse Western blot. L’évaluation de la résistance des plantes transgéniques dans des conditions de serre a montré que l'incidence de la maladie et les symptômes ont été réduits pour R. solani.
Abstract
In the present study, the potato cultivar ‘Desirée’ was transformed via Agrobacterium tumefaciens strain LBA4404 containing the plasmid pBIN19 which harbors the Ribosome Inactivating Protein (rip30). The potato leaf discs were used as an explant for transformation. The in vitro regeneration parameters (percentage of callus regenerated, number of shoots per callus, percentage of regenerated roots and percentage of the transgenic plants) were evaluated. The PCR technique was used for identification of transformed plants. Southern and Western blot analyses were applied for molecular characterization of the transgenic clones. A greenhouse assay was carried out to evaluate the resistance to Rhizoctonia solani pathogen of transgenic clones expressing the rip30 gene. The results revealed that not all the plants developed in selective medium were positive for the corresponding gene using the PCR technique. Southern blot analysis demonstrated that the tested transgenic plants integrated three copies of rip30 gene into their genome. The expression of the RIP30 protein was confirmed in the leaf extracts of the transgenic clones by Western blot analysis. Resistance evaluation of the transgenic plants in greenhouse conditions showed that disease incidence and severity were reduced for R. solani.
Tabla de contenidos
1. Introduction
1Potato (Solanum tuberosum L.) is one of the most important crops worldwide. Unfortunately, commercial cultivars are particularly susceptible to fungal and bacterial diseases leading to considerable losses in yield and quality of products (Walter et al., 2001; Khan et al., 2008). Rhizoctonia solani, the most widely spread species of Rhizoctonia, is a soil borne fungal pathogen. The most common symptom on potato result in two appearances of the disease, namely stem canker and black scurf. These are recognized as necrotic lesions on underground plant parts, and sclerotia covering progeny tubers, respectively (Carling et al., 1986).
2Plant disease control based mainly on chemical pesticides is not always effective and is commonly expensive. Furthermore, this treatment involves residual toxicity and could contribute to the selection of fungicide-resistant pathogens. Consequently, constitutive transgenic expression of genes encoding antimicrobial proteins with broad spectrum into commercial cultivars constitutes an interesting approach to reduce losses caused by pathogens.
3As a counter-defence, plants have evolved some potent defense mechanisms, including the synthesis of low-molecular weight peptides with antifungal activity (Selitrennikoff, 2001). Ribosome-inactivating proteins (RIPs) are plant enzymes that have 28 S rRNA N-glycosidase activity, which depending on their specificity, can inactivate foreign ribosomes, thereby shutting down protein synthesis. The most common cytosolic type I RIP from the endosperm of cereal grains does not act on plant ribosomes but can affect foreign ribosomes, such as those of fungi (Stirpe et al., 1992; Hartley et al., 1996).
4Some transgenic plants harboring RIP genes have already been produced (Jach et al., 1995; Bieri et al., 2000). Expression of barley seed RIP reduced development of R. solani in transgenic tobacco (Logeman et al., 1992) but had little effect on Blumeria graminis in transgenic wheat (Bieri et al., 2000). In previous study, the RIP was targeted to the apoplastic space and may exhibit a lower activity against development of the intracellular haustoria of the mildew pathogen (Punja, 2001).
5In the present paper, a transformation of the potato cultivar ‘Desirée’ is carried out for further study of the effectiveness of RIP gene in the defense process against fungal infection of R. solani.
2. Materials and methods
2.1. Plant material
6Virus free potato tubers of the tetraploid cultivar ‘Desirée’ were used for transformation. Etiolated sprouts from these tubers were surface-disinfected and single node segments were excised and placed on MS medium (Murashige and Skoog, 1962) supplemented with 100 g·l-1 of inositol, 30 g·l-1 of sucrose, 0.7% (W/V) of agar, the pH was adjusted to 5.6. Stems developing from these nodes were propagated in vitro by subculturing the top shoots or stem segments including axillary buds every 3-4 weeks. The shoots were grown at 23 ºC during a photoperiod of 16 h under a 3,000 lux light intensity.
2.2. Bacterial growth
7The disarmed Agrobacterium tumefaciens strain LBA4404, harboring the pBIN19 (13758 bp) was used. The pBIN19, kindly provided by Dr. J.I. Ruiz de Galarreta (NEIKER, Vitoria), contains the rip30 gene of 1031 bp coding for Ribosome-Inactivating Proteins of barley (Hordeum vulgare L.) (Logeman et al., 1992) and driven by CFDV promoter (Randles et al., 1992) and containing the nptII gene, coding for Neomycin Phosphotransferase II, conferred kanamycin resistance, as a selectable marker of the transgenic plants (Figure 1).
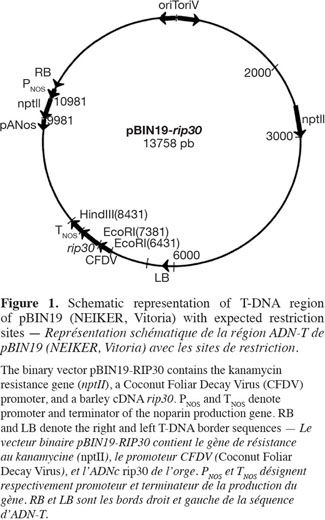
8Agrobacterium tumefaciens was grown in Luria-Bertani (LB) medium at 28 ºC. For leaf infection, bacteria cells were grown overnight at 28 °C with constant shaking (200 rpm) in YEB medium (Hooykaas et al., 1979) supplemented with 50 mg·l-1 of kanamycin, 100 mg·l-1 of streptomycin and 25 mg·l-1 of rifampycin. When OD600nm reached 0.8-1, the culture was diluted 1:10 in MS medium. The bacterial cells were collected by centrifugation and resuspended in a MS medium supplemented with 50 mg·l-1 of sucrose, pH 5.6
2.3. Plant transformation
9The leaf disc transformation method was used. Five mm leaf discs were cut from in vitro plants. Forty explants were used. The explants were infected with 30 ml of A. tumefaciens and placed at 25 °C in dark conditions for 1 h. Infected explants were co-cultivated, in the dark, for 2 days on a MS basal medium salts supplemented with 100 g·l-1 of inositol, 30 g·l-1 of sucrose, 0.7% (W/V) of agar, the pH was adjusted to 5.6 and without antibiotics to allow the bacteria to transform leaf cells. Subsequently, the leaf discs were washed with MS basal medium containing 500 mg·l-1 of cefotaxime to eliminate bacteria. After drying on sterile filter paper, they were transferred to Petri dishes containing 25 ml of the MS basal medium supplemented with 2 mg·l-1 of zeatin, 20 µg·l-1 of gibberellic acid, 20 µg·l-1 of α-naphthalene acetic acid (α-NAA), 500 mg·l-1 of cefotaxime and 100 mg·l-1 of kanamycin. They were maintained on this medium for 5 weeks at 23 °C during a photoperiod of 16 h for callus induction and shoot regeneration. The resulting shoots of 5 mm tall were regenerated during 4-5 weeks after transformation and were then individually transferred to MS medium containing kanamycin (100 mg·l-1) and cefotaxime (500 mg·l-1) for rooting. Transgenic plants were further transferred to greenhouse conditions.
10In this experiment several parameters were evaluated: percentage of induced callus, percentage of regenerated calli, number of shoots per callus, percentage of regenerated roots and percentage of the transgenic plants.
2.4. PCR amplification
11To detect rip30 gene by PCR, genomic DNA was extracted from leaf material according to Edwards et al. (1991). DNA fragment, containing the rip30 gene, of 500 bp was amplified by PCR using specific primers. The complete sequence of these primers is as follows:
12rip30: 5’CAACCCGGCGCACTTCTC3’,
13reverse rip30: 5’GGCCTTCATCTCATTGCCG3’.
14PCR amplification reactions consisted of an initial denaturation at 94 ºC for 2 min followed by 40 cycles of denaturation at 94 ºC for 1 min, hybridization at 55 °C for 1 min, extension at 72 ºC for 2 min and a post-extension at 72 ºC for 10 min. Taq DNA polymerase was used in all PCR reactions. The resulting PCR products were separated by Agarose gel electrophoresis 1% (w/v).
2.5. Southern blot analysis
15To detect the integration of rip30 gene in transgenic plants, total DNA was extracted from young leaves. Ten µg of DNA from each sample were digested with EcoRI, separated on a 0.9% agarose gel, transferred to a nylon membrane (Hybond N+, Amersham Pharmacia Biotech) and hybridized with the DIG-labeled probe of the rip30 gene. The fluorescent DNA probe was isolated from the plasmid and digested by HindIII. Hybridization, washing and detection were performed using DIG Easy Hyb (hybridization solution) and DIG Wash and Block Buffer set following the supplier’s instructions (Boehringer Mannheim).
2.6. Western blot analysis
16To evaluate the expression of the integrated rip30 gene in the genomic DNA of transgenic plants, the total soluble proteins were extracted from 300 mg of leaf tissue in 100 µg of loading buffer 3X (Laemmli, 1970). The homogenized samples were boiled for 3 min and extracts were centrifuged at 13,000 rpm for 2 min. Then 40 µl of soluble proteins were loaded onto 12% SDS-polyacrylamide gel (SDS-PAGE) according to Laemmli method (1970) and electroblotted onto a polyvinylidine difluoride (PVDF) membrane (Amersham). Immunodetection was performed using polyclonal anti-serum raised in rabbit against the RIP30 protein and goat-anti-rabbit IgG (Amersham) conjugated to horseradish peroxidise (HRP) as secondary antibody.
2.7. Greenhouse assay of the transgenic plants inoculated with R. solani
17Seven clones with confirmed insertion of the putative genes were chosen and micropropagated in vitro. After 4-5 weeks, five plants for each clone and control were transplanted into individual pots and placed in the greenhouse for further screening for resistance to the fungus Rhizoctonia solani. Strain Ci96 was provided by “Servicio de Semillas y Plantas de Vivero de Vitoria, Spain” and was grown on Potato Dextrose Agar (PDA) medium (20% [w/v] potato, 1.5% [w/v] glucose and 1.5% [w/v] agar) at 25 °C with light for 5 days, and subcultured as needed. The inoculum of isolate Ci96 was grown in 250 ml flasks containing 100 g of barley and wheat grains, and 120 ml of distilled water. Each flask was inoculated with 5 plugs of 5 mm diameter of the fungus taken from the margins of a 1 week-old culture of isolate Ci96, grown on PDA medium. Flasks were incubated at 25 °C for 18 days. Inoculum was added to pots filled with sterilized potting mixture as described by Chand et al. (1982).
18After 4 months, the crop was harvested and tubers symptoms were assessed. The symptoms evaluation was the appearance of black or brown sclerotia on the tuber surface, using the ADAS scale (Anonymous, 1976), which ranges from 0 to 25% according to the infected surfaces covered by sclerotia.
19For the statistical analysis, the ANOVA was performed to evaluate the significance of the differences in the resistance level among the clones. Means were separated using the LSD test (P < 0.05). The greenhouse assay was carried out three times into independent assays.
3. Results
3.1. Transformation and characterization of transgenic plants
20The results of transformation with the pBIN19 are indicated in table 1. Not all the plants growing on the kanamycin-containing medium were positive in the identification of the corresponding gene present on the T-DNA using the PCR technique (Table 1). PCR analysis was carried out for all kanamycin resistant plants. This analysis showed that only 47% of the plants, rooted in kanamycin medium, displayed a 500 bp amplification fragment for rip30 gene. The specific primers did not amplify the corresponding fragments in the untransformed samples (Figure 2a).

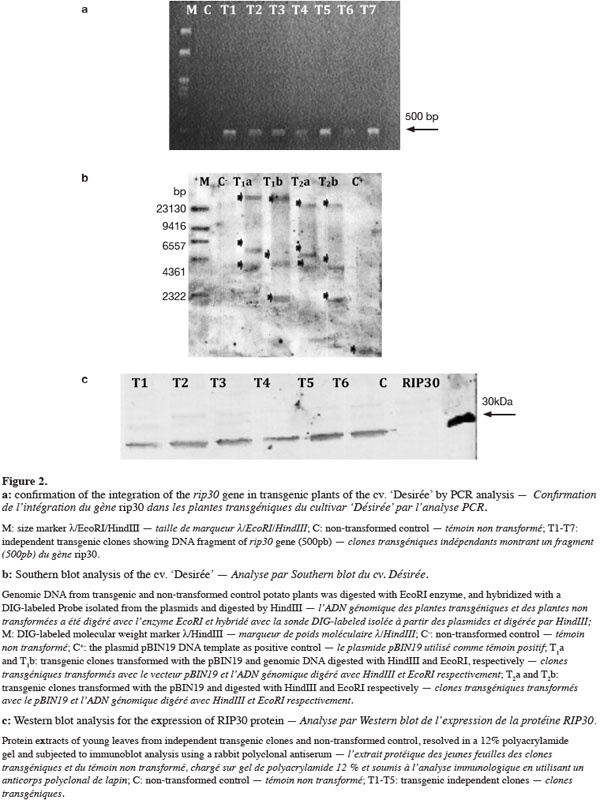
21Southern blot analysis showed that the rip30 gene was integrated into the genome of the tested transformed plants. In these transformants, three hybridizing bands were detected (Figure 2b). Southern blot is given for two selected positive PCR clone, T1 and T2, hybridized with the probes from rip30; a three bands hybridization is obtained, indicating that there could be three copies of the rip30 gene inserted for this transgenic clones. The size of the bands differed, indicating that these transformants have three copies of the rip30 gene integrated at random sites in the genome. The pattern of integration was variable. No transgene insertion was detected in non-transformed controls.
22The Western blot analysis demonstrated that the rip30 gene is transcribed and translated into proteins in the tested transgenic clones. The 30 KDa peptide of the RIP30 protein was detected as a single band in the total protein extracts from the transgenic plants with varying levels of expression in different clones (Figure 2c). The control plant non-transformed did not express the RIP30 proteins.
3.2. Resistance evaluation to the virulent isolate of R. solani in transgenic plants
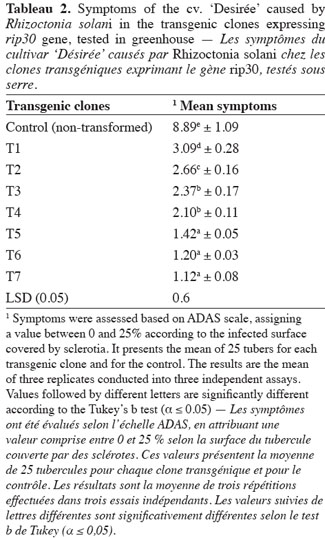
23The mean symptoms varied between the clones, carrying the rip30 gene, and ranged from 1.12 to 3.09 (Table 2). All the tested clones showed lesser symptoms than the non-transgenic control (8.89). The clone T7 showed low symptoms (1.12) revealed by a very mild black scurf symptoms. Statistically, significant differences were observed in symptom severity among the clones, which showed a reduced severity of the symptoms caused by the pathogen. T5, T6 and T7 were the most resistant clones.
4. Discussion
24In this study the potato cultivar ‘Desirée’ was transformed under experimental conditions with rip30 gene isolated from Hordeum vulgare L. The transformation was carried out via A. tumefaciens, which has been commonly used for potato transformation and plays a significant role in determining the efficiency of infection (Beaujean et al., 1998; Ducreux et al., 2005; Banerjee et al., 2006).
25Not all the plants grown on selective medium with kanamycin were positive in PCR, as reported by Kumar et al. (1995) who observed many escapees despite the selective medium. This could be explained by the fact that some plants could integrate only the gene of resistance to the antibiotic, as the transfer of T-DNA occurs from the right to the left border (Wang et al., 1984). Indeed, in the binary vectors that we used, the nptII gene conferring resistance to kanamycin is nearest the right border than rip30 gene.
26The transgenic clones (positive in PCR) were confirmed by Southern blot analysis. This showed integration of three copies of the rip30 gene into their genome. In previous reports it was shown that the variation in the number of copies is not correlated with the differences in expression level and it is due to the effect of the T-DNA position in different localization of the genome (Deblaere et al., 1987).
27The ability of the introduced rip30 gene to enhance resistance in the transgenic potato plants was studied in greenhouse conditions. The transgenic clones expressing the rip30 gene were able to reduce symptom severity of R. solani in an in vivo assay.
28These transgenic clones were also evaluated for resistance to Phytophthora infestans and no differences were found between the transgenic plants expressing the rip30 gene and the untransformed control (data not shown). These transgenic clones failed to reduce incidence and severity of P. infestans. These results confirmed that the enzyme used in our experiments did not induce resistance to several fungi, and showed a narrow spectrum of antifungal activity. This could be explained by a weak antifungal activity of this enzyme (Lorito et al., 1998).
29The research presented here demonstrates a successful transformation of potato plants and the results revealed that the rip30 gene was successfully integrated into the genome of transgenic potato and expressed by producing the RIP30 protein. The transgenic clones obtained reduced incidence and severity of R. solani but failed to enhance resistance to P. infestans.
Bibliographie
Anonymous, 1976. Manual of plant growth stages and disease assessment keys. Harpenden, Herts, UK: Plant Pathology Laboratory, Ministry of Agriculture, Fisheries and Food.
Banerjee A.K., Prat S. & Hannapel H.J., 2006. Efficient production of transgenic potato (S. tuberosum L. ssp. andigena) plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci., 170, 732-738.
Beaujean A., Sangwan R.S., Lecardonnel A. & Sangwan-Norreel B., 1998. Agrobacterium-mediated transformation of three economically important potato cultivars using sliced intermodal explants: an efficient protocol of transformation. J. Exp. Bot., 49, 1589-1595.
Bieri S., Potrykus I. & Futterer J., 2000. Expression of active barley seed ribosome-inactivating protein in transgenic wheat. Theor. Appl. Genet., 100, 755-763.
Carling D.E. & Leiner R.H., 1986. Isolation and characterization of Rhizoctonia solani and binucleate R. solani-like fungi from aerial stems and subterranean organs of potato plants. Phytopathology, 76, 725-729.
Chand T. & Logan C., 1982. Reaction of ten potato cultivars to stem canker and black scurf of potato caused by Rhizoctonia solani. Ann. Appl. Biol., 100, 102-103.
Deblaere R. et al., 1987. Vectors for cloning in plant cells. In: Wu R. & Grossman L., eds. Methods in enzymology. New York, USA: Academic Press, 277-292.
Ducreux L.J.M., Morris W.L., Taylor M.A. & Millam S., 2005. Agrobacterium-mediated transformation of Solanum phureja. Plant Cell Rep., 24, 10-14.
Edwards K., Johnstone C. & Thompson C., 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res., 19, 1349.
Hartley M.R., Chaddock J.A. & Bonness M.S., 1996. The structure and function of ribosome inactivating proteins. Trends Plant Sci., 1, 254-260.
Hooykaas P.J.J., Roobol C. & Schilperoort R.A., 1979. Regulation of the transfer of Ti-plasmids of Agrobacterium tumefaciens. J. Gen. Microbiol., 110, 99-109.
Jach G. et al., 1995. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J., 8, 97-109.
Khan R.S., Sjahril R., Nakamura I. & Mii M., 2008. Production of transgenic potato exhibiting enhanced resistance to fungal infections and herbicide applications. Plant Biotechnol. Rep., 2, 13-20.
Kumar A. et al., 1995. Agrobacterium-mediated transformation of five wild Solanum species using in vitro microtubersin. Plant Cell Rep., 14, 324-328.
Laemmli U.K., 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685.
Logeman J. et al., 1992. Expression of a barley ribosome inactivating protein leads to increased fungal protection in transgenic tobacco plants. Biotechnology, 10, 305-308.
Lorito M. et al., 1998. Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc. Natl Acad. Sci. USA., 95, 7860-7865.
Murashige T.H. & Skoog F., 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol., 15, 473-497.
Punja Z.K., 2001. Genetic engineering of plants to enhance resistance to fungal pathogens-a review of progress and future prospects. Can. J. Plant Pathol., 23, 216-235.
Randles J.W. et al., 1992. Localisation of coconut foliar decay virus in coconut palm. Ann. Appl. Biol., 121(3), 601-617.
Selitrennikoff P., 2001. Antifungal proteins. Appl. Environ. Microbiol., 67, 2883-2894.
Stirpe F. et al., 1992. Ribosome-inactivating proteins from plants: present status and future prospects. Biotechnology, 10, 405-412.
Walter R.S., Rosemary L., Gary F.D. & Weingartner D.P., 2001. Compendium of potato diseases. St Paul, MN, USA: American Phytopathological Society.
Wang K., Herrera-Estrella L., Van Montagu M. & Zambryskie P., 1984. Right 25 bp terminus sequence of the nopaline T-DNA is essential for and determines direction of DNA transfer from Agrobacterium to the plant genome. Cell, 38, 455-462.
Para citar este artículo
Acerca de: Mahmoud M’Hamdi
Higher Institute of Agronomy of Chott-Mariem. BP. 47. TN-4042 Sousse (Tunisie). E-mail: mahmoud.mhamdi@iresa.agrinet.tn
Acerca de: Hela Chikh-Rouhou
Higher Institute of Agronomy of Chott-Mariem. BP. 47. TN-4042 Sousse (Tunisie).
Acerca de: Naima Boughalleb
Higher Institute of Agronomy of Chott-Mariem. BP. 47. TN-4042 Sousse (Tunisie).
Acerca de: José Ignacio Ruiz de Galarreta
Neiker Basque Institute for Agricultural Research. PO. Box 46. E-01080 Vitoria (Spain).






