- Home
- Volume 19 (2015)
- numéro 4
- Molecular diversity of Cuban cassava (Manihot esculenta Crantz) cultivars assessed by simple sequences repeats (SSR)
View(s): 0 (0 ULiège)
Download(s): 0 (0 ULiège)
Molecular diversity of Cuban cassava (Manihot esculenta Crantz) cultivars assessed by simple sequences repeats (SSR)

Editor's Notes
Received on September 9, 2013; accepted on June 29, 2015
Résumé
Diversité moléculaire de cultivars cubains de manioc (Manihot esculenta Crantz) analysés par séquences simples répétées (SSR)
Un total de 36 marqueurs microsatellites (SSR) ont été utilisés pour analyser la diversité génétique de 163 accessions cultivées de manioc (Manihot esculenta Crantz), 94 d'entre elles à partir de la collection de matériel génétique de manioc cubain et 69 autres génotypes de différents pays d'Afrique, d’Asie et d’Amérique latine, conservés au Centre International d'Agriculture Tropicale (Colombie). L'étude a été menée afin de déterminer la diversité génétique au sein et entre toutes les accessions pour promouvoir une meilleure conservation et utilisation. Trente-quatre SSR étaient polymorphes et utiles dans les études sur la diversité génétique. Les cultivars cubains ont montré le plus grand nombre de loci avec en moyenne 5,8 allèles et 100 % étaient des loci polymorphes, comme ceux du Guatemala. L'hétérozygotie moyenne observée (HO) était élevée (0,5918 ± 0,0351), avec les valeurs les plus élevées de HO pour les génotypes de Cuba (0, 6016) et de Tanzanie (0,6459). L'hétérozygotie globale (HT) était également élevée (0,6538 ± 0,1770), mais seulement 7,4 % (GST = 0,0740 ± 0,0377) était dû à des différences entre les cinq pays étudiés. Les coefficients de la différenciation génétique (estimé par F-Statistics) étaient bas à modérés et 17 allèles uniques ont été trouvés avec une faible fréquence d'occurrence entre les génotypes cubains. Les résultats fournissent la première caractérisation moléculaire des génotypes de manioc conservés à Cuba et confirment leur large variabilité génétique. L’exploitation de cette information importante contribue à améliorer les stratégies nationales de conservation et d'utilisation du patrimoine génétique, en particulier dans les programmes d'amélioration génétique du manioc.
Abstract
A total of 36 microsatellites (SSR) markers were used to analyze the genetic diversity of 163 accessions of cultivated cassava (Manihot esculenta Crantz), 94 accessions of them from the Cuban Cassava Germplasm Collection and 69 genotypes from different countries and conserved at the International Center for Tropical Agriculture (Colombia). This study was carried out to determine genetic diversity within and between all accessions to promote their better use and conservation strategies. Thirty-four of those markers were used for the genetic diversity study based on their higher polymorphism. The Cuban cultivars showed the highest average allele number per loci with 5.8 and 100% of the loci were polymorphic, as well as those from Guatemala. The average proportion of individual heterozygocity observed (HO) was high (0.5918 ± 0.0351), while the highest HO rates were observed in groups of genotypes from Cuba (0.6016) and Tanzania (0.6459). The total heterozygocity (HT) was high (0.6538 ± 0.1770), but only 7.4% (GST = 0.0740 ± 0.0377) was due to differences between the five countries studied. Genetic differentiation coefficients (estimated by F-statistics) were low to moderate (FST > 0.04) and 17 unique alleles with low frequency were found in Cuban cultivars. The results provide the first molecular characterization of Cuban cassava genotypes and showed a wide diversity among landraces from Cuba. Application of this valuable information can be used for genetic diversity conservation and genotype identification studies for the genetic breeding program of cassava.
Table of content
1. Introduction
1Cassava (Manihot esculenta Crantz) is an important crop that is commonly grown in tropical and subtropical areas. It grows very well in a less fertile soil in contrast to many other crops, even in marginal areas and it is able to express a good yield of roots, under favorable production conditions or not; millions of people around the world depend on it to subsist (Vásquez et al., 2012). However, its potentialities are far from being adequately exploited and knowledge of its genetic diversity is limited.
2The application of advanced molecular breeding technologies could be useful for cassava research studies. This has been demonstrated through research on worldwide important crops (Raji et al., 2009). Molecular marker technologies can assist conventional breeding efforts as valuable tools for the analysis of genetic relatedness, the identification and selection of desirable genotypes for crosses as well as for germplasm conservation.
3Molecular genetic markers can be used to map genomes, identify regions of the genome controlling a trait, and follow a segment of interest of the genome in a plant breeding scheme (Kunkeaw et al., 2010; Bang et al., 2011; Ferguson et al., 2012). Various research studies on cassava genetic variation were based on molecular markers such as Restriction Fragment Length Polymorphisms (RFLP) (Fregene et al., 1997) or Random Amplified Polymorphic DNA (RAPD) (Vieira et al., 2011). Nevertheless, Simple Sequence Repeats (SSRs) or microsatellites are particularly attractive for diversity studies (Siqueira et al., 2009; Turyagyenda et al., 2012) because they are abundant in plants and they have a high level of polymorphism (Mba et al., 2001).
4Microsatellites are powerful markers for both genetic diversity studies and marker assisted selection (MAS) and they have been used in many plant species (Roubus et al., 2010; Lekha et al., 2011). In addition, SSRs markers were used to evaluate the molecular diversity of some cassava germplasm (Raghu et al., 2007; Kunkeaw et al., 2011; Montero-Rojas et al., 2011) or to compare genetic diversity between improved cassava cultivars and landraces from Africa, Latin America, Asia and four Manihot wild species (Whankaew et al., 2012).
5In Cuba, cassava serves as one of the main sources of energy-rich food for humans and animals. The “Instituto de Investigaciones de Viandas Tropicales” (INIVIT) preserves an important germplasm collection with 540 cassava genotypes, from different collects made in Cuba (nationwide), some of them are improved cultivars and others were introduced in Cuba from other countries by germplasm’s exchange. Previous morphological and agronomic characterization revealed a wide genetic variability (Milián et al., 2000). Current advance in the development of methods using DNA polymorphisms as molecular markers provides alternative methods for landraces characterization and especially, microsatellites could help to reveal or to evaluate the genetic variability in the Cuban germplasm.
6Genetic diversity of cassava cultivars was assessed in 94 Cuban accessions from the germplasm held at INIVIT, as a mean of designing better conservation approaches and identifying progenitors with a wide genetic base for breeding. Therefore, the objectives of this study are:
7– to assess the genetic diversity of 94 Cuban cassava accessions with an appropriate number of SSR markers;
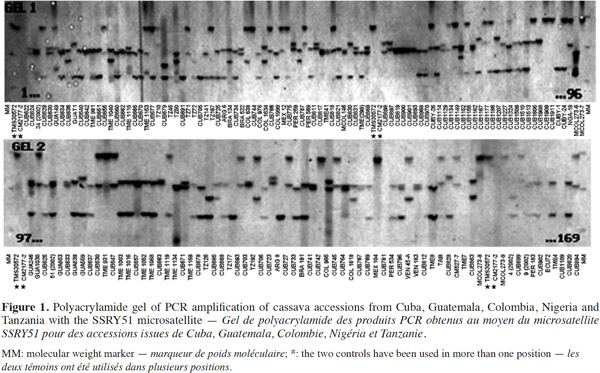
8– to determine genetic relationships between Cuban accessions and other 69 genotypes from different geographic origin.
2. Materials and methods
2.1. Plant material
9A total of 163 cassava accessions from different geographical origin were used (Table 1), 94 of them are Cuban (collected or obtained by breeding programs in Cuba) and were selected from the Cuban cassava germplasm collection held at “Instituto de Investigaciones de Viandas Tropicales” (INIVIT); these were selected according to their genetic or productive potential according to previous morphoagronomic studies (Milián et al., 2000), and breeders and producers criteria. The remaining 69 accessions were chosen from the International Centre of Tropical Agriculture (CIAT) germplasm collection in Colombia; these accessions represent the main cassava producing countries. These genotypes were used before in similar studies of diversity with the same SSR set we used in this investigation (Fregene et al., 2003). The genotypes 'TMS30572' and 'CM2177-2' were used as controls because of their very well-known genetic pattern for the evaluated microsatellites.
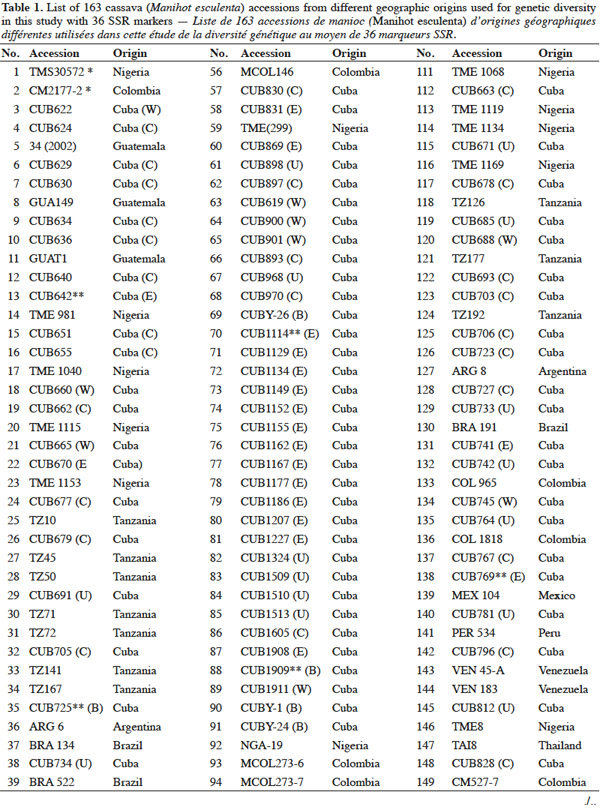
10Woody stakes of each cultivar, 20 to 30 cm length, were harvested from all Cuban genotypes and planted in a greenhouse, the young leaf tissue from the buds generated from cuttings were used to obtain the genomic DNA for each Cuban accession. The DNA from the others 69 accessions was kindly given by the Cassava Breeding Program of CIAT.
2.2. DNA extraction
11Two hundred mg of young and healthy leaves from each Cuban accession, collected from small buds grown under greenhouse conditions, were used. The total genomic DNA of each cultivar was carried out according to Dellaporta et al. (1983). Quality of total DNA was checked by electrophoresis in 0.8% agarose gel (Amresco, Solon, OH, USA) using 0.55 M Tris-EDTA buffer. Each DNA was visualized by staining in 0.5μg·mg-1 Ethidium bromide; dilutions of phage lambda DNA of known concentration were used as control. Finally, DNA obtained from each accession was quantified by fluorometry (DyNA Quant TM. 200 Fluorometer, Hoefer Pharmacia Biotech).
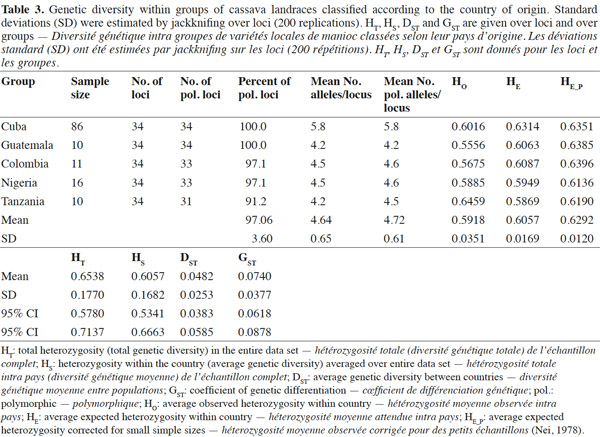
2.3. Polymerase Chain Reaction (PCR) with SSR markers
12A dilution of each DNA sample was done to a final concentration of 10 ng·μl-1 and was used for PCR analysis. The cocktail for PCR was prepared as follows: 50 ng DNA, 0.2 mM dNTPs, 2.5 buffering solution 10 X, 2 mM MgCl2, 1.25 μM primer F, 1 U of DNA polymerase Taq and H2O (quality HPLC) until 25 μl final volume. Amplification was carried out in a thermocycler PTC-100, the thermocycling profile consisted of an initial denaturation step for 1 min at 94 °C, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C or 45 °C for 1 min and primer extension at 72 °C for 1 min, and a final extension cycle of 5 min at 72 °C. The PCR product stayed at 4 °C during an indefinite time. The PCR product was denatured and electrophoresed on 4% polyacrylamide gels, using an automated DNA sequencer ABI model 377 (Pelkin Elmer Inc.).
13Gels were fixed in 10% HAc and the bands were revealed by staining with silver nitrate (SilverSequence TM DNA Sequencing System, Promega) according to Fregene et al. (2002).
14The raw gel data were extracted using the GENESCAN package of ABI PRISM for Windows and GENOTYPER software (Pelkin Elmer Inc.). The extracted data were exported as allele sizes to Microsoft Excel (Microsoft Inc.) for further formatting as input files for statistical analysis. A strictly diallelic model of inheritance was applied; in other words, markers having three or more alleles were eliminated according to Fregene et al. (2003).
2.4. Gene diversity and genetic differentiation studies
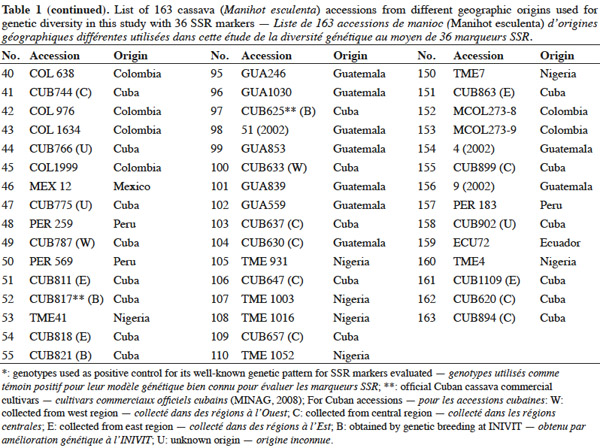
15SSR markers used in this study are listed in table 2 and were supplied by CIAT, and consisted of 36 primer pairs, which were selected from a subset of SSR markers developed at CIAT (Mba et al., 2001). The 36 primer pairs were selected based on being polymorphic, having clear reproducible allele patterns, high polymorphic information content (PIC), evenly spread across the cassava genome (Mba et al., 2001; Hurtado et al., 2008).
16Gene diversity and genetic differentiation analyses were carried out with data from the polymorphic SSR markers that showed distinct and scorable DNA bands, their diallelic nature and clear patterns.
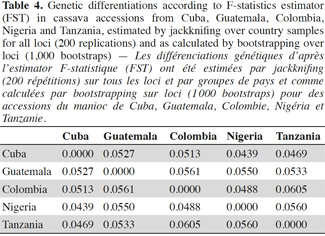
17All unambiguous SSR fragments from each of the polymorphic SSR markers were scored using binary numbers as presence (1) and absence (0) of an allele across all 163 genotypes. The genotypes TAI8 (Thailand) and ECU72 (Ecuador) were also eliminated because of too many missing data. From the obtained matrix, similarity degree within and between countries were calculated using Dice similarity coefficient (Dice, 1945). The following analysis related with diversity and differentiation studies only included the information related with cultivars from the most widely represented countries in this investigation: Cuba, Colombia, Guatemala, Nigeria and Tanzania (hereinafter, consider all accessions from each country as a ‘group’ or ‘population’).
18Genetic diversity within and between countries was estimated using the software package GENSURVEY (Vekemans et al., 1997), with the following statistics: percentage of polymorphic loci, mean number of alleles per polymorphic locus, average observed heterozygosity (HO) and average gene diversity (HE) (Nei, 1978). For all loci and for all accessions, total heterozygosity (HT) and the proportion of among-accession differentiation (GST) were estimated according to Nei (1978). Standard deviations for the above parameters were estimated over loci and sampled by jackknifing (Quenoille, 1956) using 200 replications. Given the small evolutionary divergence times for the accessions, the infinite allele model (Kimura et al., 1964) was assumed.
19Genetic differentiation was quantified by the F-statistic estimator (FST, theta) (Wright, 1951), as described by Weir et al. (1984), using FSTAT 2.9 (Goudet, 2001), and by GST (Nei, 1978); FSTAT performs bootstrapping over loci and, given the large number of unlinked SSR loci employed in this study, it provides rigorous testing of hypotheses of genetic differentiation. FST gives a similar estimate of genetic differentiation as GST but GST takes into account variation in samples (accessions groups = accessions group by country) sizes, which is relevant in this study. FST values were estimated per allele, per locus and overall. Confidence intervals were calculated per locus over samples, and over loci by jackknifing (200 replicates), and by bootstrapping (1,000 bootstraps), over loci.
20Pairwise genetic distances between individual accessions were calculated from the raw allele size-data based on the 1-proportion of shared alleles (1-PSA) (Bowcock et al., 1994), and estimated using the computer program “MICROSAT” (Minch, 1996). Distances based on 1-PSA give a more exact representation of genetic relationships when using closely related genotypes and SSR markers, compared with other distance estimates (Bertin et al., 2001). The distance matrix generated was subjected to principal component analysis (PCA), using the program JMP (SAS Institute, 1999) with the purpose to obtain a structure of relationships among cassava accessions; a scatter plot of all cassava accessions and, another independent scatter plot of only Cuban accessions in two dimensions were produced.
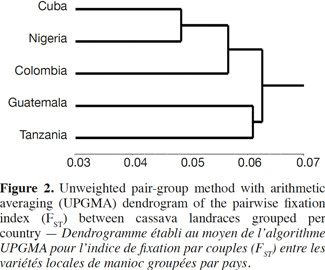
21A comparison matrix between countries was created with FST values using FSTAT (Goudet, 2001), and the relationship was analyzed by cluster analysis, using the unweighted pair group method using arithmetic averages (UPGMA). UPGMA clustering was performed using the SAHN programme of NTSYS-PC (Rohlf, 1998) to construct the dendrogram with a view to analyze the relationship between the Cuban landraces and the other cultivars by selecting the tree plot option in the same software package.
3. Results and discussion
22Genetic diversity in cassava was evaluated in 163 genotypes from different geographic origins with 36 SSR primers to obtain a quantitative estimate of diversity in local cassava varieties; only two primers were monomorphic (SSRY-127 and SSRY-132) and they were excluded of the final analysis. Thirty-four polymorphic SSR primers that produced clear and scorable bands were analyzed.
23The results confirmed the high polymorphism of SSR markers and the figure 1 shows the observed polymorphism with the primer SSRY51 in the studied accessions from the five countries with the highest number of evaluated genotypes.
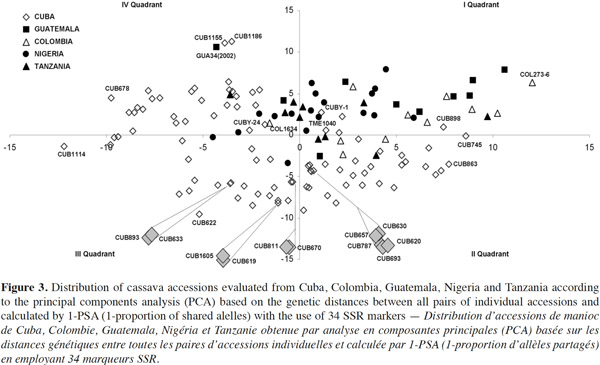
24Data from the genetic distance analysis based on 34 polymorphic microsatellites were sufficient to distinguish all genotypes tested, and all images of the electrophoretic separations for those SSRs are available on the following web site: http://isa.ciat.cgiar.org/molcas/estudios.jsp?code=7&pais=Cuba. All those markers have shown adequate values in their polymorphism rates (PIC) in several previous diversity studies with other cassava genotypes (Mba et al., 2001; Hurtado et al., 2008; Lekha et al., 2011).
25The number of SSR primer loci detected among all cassava landraces ranged from 2 to 10 alleles per locus with a 4.64 average, which is comparable with the values obtained by Raji et al. (2009) in a research on African cassava cultivars and elite germplasm from Asia and Latin America. However, Siqueira et al. (2009) only found an average of 3.3 alleles per locus in a similar genetic diversity study between Brazilian cassava landraces assessed with the same SSRs markers. Kawuki et al. (2009), working with three different populations, from Asian, African and American cassava reported numbers of alleles ranging between 3 and 11.
26Seventeen unique alleles with low appearance frequency (in all cases less than 25%) were found in Cuban accessions, in 12 of the 34 polymorphic markers evaluated: SSRY4 (0.04), SSRY20 (0.006), SSRY38 (0.005), SSRY59 (0.006 and 0.079), SSRY63 (0.033), SSRY69 (0.023), SSRY100 (0.011), SSRY103 (0.052, 0.012, 0.012 and 0.006), SSRY135 (0.005), SSRY151 (0.05), SSRY171 (0.012 and 0.036) and SSRY177 (0.014). Unique alleles were also found in genotypes from Colombia (6), Nigeria (3), Tanzania (3) and Guatemala (1).
27The high polymorphism showed for this primers set also confirms the appropriateness of SSR markers for use in cassava genome analysis (Mba et al., 2001). The utility of SSR has been widely confirmed by recent studies on cassava diversity (Hurtado et al., 2008; Kunkeaw et al., 2010; Lekha et al., 2011).
28The unique and broad diversity of cassava landraces found in Cuba reveals an invaluable germplasm resource for cassava breeding targeted to the region. The unique diversity suggests that the Cuban germplasm might have particular genetic information that will be more widely studied in the future.
3.1. Estimation of genetic diversity
29In the genetic diversity indices calculated from the SSR data within and between countries, a high genetic polymorphism was observed for all microsatellites tested. On an average, there were more than 98% of polymorphic loci across all accessions and countries (see http://isa.ciat.cgiar.org/molcas/estudios.jsp?code=7&pais=Cuba). As the best represented countries in this research were Cuba, Colombia, Guatemala, Nigeria and Tanzania, hereinafter other studies were developed to deepen into their diversity and genetic differentiation.
30Cuba and Guatemala genotypes showed 100% of polymorphic loci (Table 3); a high number of alleles was found per locus, with an average of 4.64 ± 0.0351; Cuban landraces revealed the highest value (5.8 alleles per polymorphic locus), followed by cultivars from Colombia and Nigeria with 4.6 alleles per locus. Cassava accessions showed a mean of 4.72 alleles per polymorphic locus across the accessions.
31Fregene et al. (2003) reported an average number of alleles of 6.0 for cassava landraces from Colombia and 5.2 for Brazilian cassava. Turyagyenda et al. (2012) found an average number of 5.923 in fifty-four cassava accessions from Uganda. A study conducted on cassava in Puerto Rico by Montero-Rojas et al. (2011) reported an average of 7.15 alleles per locus ranging between 2 and 14 alleles per locus. Therefore, the number of alleles among cassava accessions from Cuba is comparable to those previous results from Colombia, Brazil and Uganda.
32The probability that two randomly selected alleles in a given accession are different, estimated by the average expected heterozygosity (HE), on average, could be considered high (0.6292 ± 0.0120), corrected for small simple sizes according to Nei (1978), with the highest HE being found for Cuban cultivars (0.6314), followed by Colombian (0.6087) and Guatemalan (0.6063) accessions.
33The average observed heterozygosity (HO) was also high (0.5918 ± 0.0351), which confirms cassava’s outcrossing and highly heterozygous nature (Fregene et al., 2003). Cultivars from Tanzania and Cuba show the highest HO indexes, 0.6459 and 0.6016, respectively.
34These results reveal a comparable level of polymorphism in all groups from Africa (Nigeria and Tanzania) and Latin America (Colombia and Guatemala), and show a comparable polymorphism level in Cuba with regard to the level found previously in other populations studied by Fregene et al. (2003). It is interesting that although the overall heterozygosity (HT) in all genotypes was high (0.6538 ± 0.1770), only 7.4% (GST = 0.0740 ± 0.0377) of this value was due to differentiation among cassava genotypes within countries, the rest is due to variations between countries (HS).
35Several studies have indicated that the number of DNA markers used for genetic studies in plants varies with the total number of accessions assessed (Thotttapilly et al., 1996). The reliability of estimates for genetic variation, such as HE, HO, FST and genetic distances, depends more on the number of loci than the number of individuals sampled (Baverstock et al., 1996). In cassava, Fregene et al. (2003) demonstrated that a minimum of 30 unlinked markers is required to obtain the maximum amount of information on allelic diversity; Hurtado et al. (2008) confirmed that 36 SSR are sufficient for cassava genetic studies.
36The results indicate an important genetic variability in the populations studied, which represents a potential that can be useful for improving national germplasm conservation strategies and especially for Cuba. This significant genetic diversity should be adequately preserved in germplasm collection and also usefully exploited in breeding programs. These results also point out the necessity to deepen into the knowledge of the genetic potentialities of the studied Cuban genotypes.
3.2. Genetic differentiation
37For genetic differentiation, pairwise calculations of FST (theta) over all loci between pairs of country accessions groups provide a picture of germplasm exchange, based on their genetic similarity coefficient, between countries. All FST (theta) estimators were low to moderate (Table 4), superior to 0.04 as estimated by jackknifing and as calculated by bootstrapping, with a 99% confidence interval (FST values ranged from 0.0439 to 0.0605). The highest FST value was reached between Colombia and Tanzania (0.0605), followed by Guatemala and Colombia (0.0561) and between Nigeria and Tanzania (0.0560).
38According to stadigraph-F calculated, the average value of FST (0.05 ± 0.006) is close to that obtained for GST (0.0427 ± 0.0385), estimated by FSTAT on all accessions from the five countries and for all loci, and indicated a differentiation from low to moderate within the five collections (countries) studied.
39On the other hand, although the overall heterozygosity (HT) considering all the genotypes was high (0.6538 ± 0.1770), only 7.4% (GST = 0.0740 ± 0.0377) of this was due to differentiation among cassava accessions within the same country, the rest is due to variations between countries (HS = 0.6057 ±0.1682). Similar results were observed by Fregene et al. (2003) and Montero-Rojas et al. (2011) during two previous genetic diversity studies with cassava accessions from different African and Latin American countries.
40Estimates of FST over all loci between pairs of countries provide an idea on germplasm flow between them, which has been limited in Cuba due to its geographic isolation.
41The pairwise FST (theta) estimates resulted in a dendrogram of the landraces that shows the relationships between African and Latin American genotypes (Figure 2). In general, a low differentiation among countries was observed or at least those accessions are as different as equals.
42The close association between cultivars from Cuba and Nigeria results to be interesting if the geographical remoteness of countries and diversity indicators and differentiation observed are considered during this investigation. This result may be due to several reasons and references found to explain them are limited. However, this is valuable information on the studied germplasm for cassava genetic improvement in Cuba.
43Two previous studies could provide some elements although in both, other accessions not included in this study were studied. Cruz et al. (2011), during a diversity study using microsatellites markers, found a close relationship between the genotypes from Cuba and Colombia in the group of accessions that presented the highest heterozygosity (0,592). Also, Whankaew et al. (2012) during the study of 33 cassava accessions from 17 countries observed similar close relationships between groups of accessions from Cuba, Nigeria and Colombia.
44During many years, common projects were developed between the cassava breeding programs from Cuba and CIAT in Colombia (S. Rodríguez, personal communication), from where every year new improved lines were delivered to be evaluated by growers in Cuba. At the same time, similar convergence points exists between CIAT and other countries from Africa (H. Ceballos, personal communication); so there is a contact point between the cultivated cassava raised by growers in all these countries because of the germplasm exchange.
45A graphical representation of the genetic relationships between all genotypes studied was obtained by a principal component analysis (PCA) performed on the genetic distance matrix. The accessions distribution based on genetic distances between all pairs of individual landraces and calculated by the 1-proportion of shared alleles (1-PSA) statistic (Bowcock et al., 1994) is showed in figure 3. The first two principal components explained 84.6% of the total variation. The genetic distance estimator 1-PSA has been found to be the most reliable for SSR data, assuming the infinite allele model (Kimura et al., 1964), and close relationships (Bertin et al., 2001).
46Principal component analysis represents graphically the relationships among accessions from different geographic areas with the peculiarity that the largest number of accessions corresponds to Cuba. This analysis showed a wide dispersion of the genotypes, evidencing a rich genetic variability, especially those coming from Cuba. The presence of some Cuban genotypes in particular positions and outside of any possible cluster is noteworthy (see e.g. CUB1114, CUB1155 and CUB1186). Most of them were collected from the east of the island and presumably share less genetic information with the other accessions studied. In general terms, this genetic structure is comparable with the one observed by Fregene et al. (2003) in which Cuban genotypes were inserted now. Well differentiated diversity groups were not observed indicating that it is very likely that dispersion is due to differential bands generated by SSR evaluated and shown in various combinations and frequencies.
47The results showed that most of studied accessions share an important part of their genetic information. In case of Cuban germplasm, the accessions were collected many years ago in production fields in several regions of the country where edaphoclimatic conditions are different. The six Cuban accessions that are farthest away from PCA distribution ends (see CUB622, CUB678, CUB745, CUB1114, CUB1155 and CUB1186) were collected in savanna soils; three were from the east region of the country (Holguín) and two from the west (Matanzas). In the center of the figure, two promising cultivars from Cuba with two representing Colombia (CIAT international collection) and Nigeria (IITA collection) (see CUBY-24 and TME1040; CUBY-24 and COL1634) are located. The four accessions are the result of national breeding schemes but CIAT has been a common point of origin.
48The principal components analysis based on genetic distances showed several Cuban accessions closely related, for example, in the second quadrant: CUB620, CUB630, CUB657, CUB693 and CUB787. The first four were collected in the municipality of Santo Domingo (Villa Clara) in savanna conditions from the central region of Cuba, while CUB787 came from Pinar del Río province in the west of the island. On this point, it is important to carry out a depth study in the future on morphological and agronomic characteristics of each.
49The lack of previous researches concerning such relation in accessions from the Cuban cassava germplasm, limits the scientific arguments that can explain this result. However, this suggests that they might possess important distinctive features from the genetic point of view, and therefore, those genotypes could have special characteristics for cassava genetic breeding.
50According to experiences of similar researches in other countries, these results may be related to the role of seedlings, human and natural selection, the heterozygous nature of cassava (Ojulong et al., 2010) and also with the influence of geographic isolation imposed by an insular environment during thousands of years.
51Ojulong et al. (2010) consider that agricultural practices of slash and burn by Amerindian farmers and the allogamous nature of cassava have been demonstrated to produce a large pool of volunteer seedlings that may be a valuable source to produce new varieties through natural and human selection. The production of new varieties not only maintains a high level of genetic diversity but also serves as insurance against crop failure due to biotic and abiotic stresses, once the adaptation to the new environmental conditions of each country or region takes place in new genotypes.
52Taking into consideration the behavior of genotypes from Cuba in comparison with the rest of African and Latin American countries studied, a principal components analysis (PCA) to assess genetic variability within all Cuban cassava accessions was conducted (Figure 4). The first two principal components account 86.2% of the total variation.
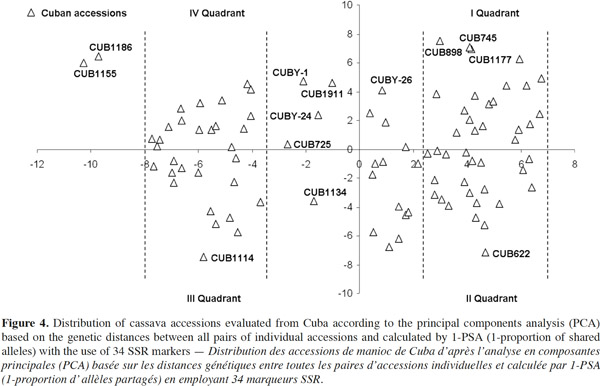
53A wide variability among Cuban accessions could be noticed but no type of grouping is clearly appreciated between them. However, a trend toward a higher concentration of accessions into two main sectors was noted.
54Interestingly, almost all accessions that were located in distant positions now and before the study with five countries (Figure 3) were collected in the east of Cuba. The eastern region shows contrasting climatic conditions such as: high temperatures, savanna and high mountains, very rainy climate or semi-desert areas. From a historical-cultural perspective, that area has been considered the territory where the first inhabitants settled in Cuba and probably the cultivated cassava in the area came, like them, from South America (Valdés et al., 1991).
55Therefore, interesting findings have been obtained in this study. Cultivars from the eastern region of the country may have distinctive features or peculiarities to differentiate them from the rest of the germplasm conserved in Cuba. Even these characteristics may be different from those found in continents. For such reasons, further researches on genetic diversity are being developed (in progress).
56Cuba is a long and narrow island where cassava has been cultivated for thousands of years (Valdés et al., 1991; Vásquez, 2010), because of this, probably the geographic isolation and limited germplasm exchange had a direct impact on the occurrence of a peculiar differentiation process in accessions coming from the east of the island.
57This finding is important and perhaps, a hypothesis could be stated considering that Cuban genotypes after many years of cultivation under insular conditions could set spontaneous deleterious mutations as a result of genetic drift in time. Fregene et al. (2003) consider that in these conditions also act the traditional cassava propagation system (vegetative or asexual), the natural and artificial selection and the allogamous nature of the crop. Likewise, cassava diversity is influenced significantly by growers during planting material exchange and in the selection and extension process of the best cultivars (Montero-Rojas et al., 2011). This is a hypothesis that, in the absence of previous studies that can be used as reference, rests on the existence of several unique and rare alleles in these accessions and suggests the convenience of further studies aimed at evaluating the specific response to adverse factors or expression of traits of interest.
58At the same time, this study confirms one important thing: the Cuban germplasm studied is an important reservoir of genetic variation that will be basic for future food security actions. There is evidence that, the level of genetic diversity is fundamental for world food security and its maintenance is considered crucial in preventing the loss of evolutionary potential and ensuring the survival of given species because of its association with fitness traits and increased biological success.
59Notably, the estimation of genetic variation in the Cuban germplasm was performed on traditional cultivars, which according to Vimala et al. (2011), are usually characterized by their dynamics in crop fields and are genetically more heterogeneous, and may also provide an excellent source of genes for adaptive traits and pest resistance.
60Recent morphological and agronomic studies on a significant group of these Cuban accessions also showed the existence of wide genetic variability in the native germplasm, as well as their potentialities for breeding and human and animal consumption and for their high yields and high dry matter percentage in roots (Beovides et al., 2013; Beovides et al., 2014).
61In general, molecular analysis applied to genotypes from the Cuban cassava germplasm, support the usefulness of this type of studies to better understand the genetic variability in cassava producing countries and in the case of Cuba, allowed to estimate parameters of genetic variation and to evaluate possible genetic relations of Cuban accessions with some other genetic relatives in America and Africa. All this contributes to better use and conservation of diversity, and generates wide useful information for possible use in better strategies for crop genetic breeding.
4. Conclusions
62Thirty-four polymorphic microsatellite markers used were useful to differentiate Cuban cassava accessions, and allowed to estimate the genetic relation between them and the evaluated genotypes from other geographical origins.
63The use of studied microsatellite markers on Cuban cassava accessions showed a high genetic differentiation between them and although their diversity was high, the relation with cultivated genotypes from Guatemala, Colombia, Nigeria and Tanzania was evident.
64Acknowledgements
65We would like to thank all of the technical staff from INIVIT and CIAT due to their contribution, interest and support shown to this project. We are very grateful to the Cassava Biotechnology Network (CBN) due to the financial support for conducting researches.
Bibliographie
Bang T., Raji A. & Ingelbrecht I., 2011. A multiplex microsatellite marker kit for diversity assessment of large cassava (Manihot esculenta Crantz) germplasm collections. Plant Mol. Biol. Rep., 29(3), 655-662.
Baverstock P.R. & Moritz C., 1996. Project design. In: Hillis D.M., Moritz C. & Mable B.K., eds. Molecular systematics. Sunderland, MA, USA: Sinauer Associates, 17-27.
Beovides Y. et al., 2013. Cultivares cubanos de yuca (Manihot esculenta Crantz) con rendimiento y potencial genético para la agroindustria. Centro Agrícola, 40(3), 71-78.
Beovides Y. et al., 2014. Caracterización morfológica y agronómica de cultivares cubanos de yuca (Manihot esculenta Crantz). Cultivos Tropicales, 35(2), 43-50.
Bertin P., Grégoire D., Massart S. & De Froimont D., 2001. Genetic diversity among European cultivated spelt revealed by microsatellites. Theor. Appl. Genet., 102, 148-156.
Bowcock A.M. et al., 1994. High resolution of human evolution with polymorphic microsatellites. Nature, 368, 455-457.
Cruz A.C. et al., 2011. Diversidad genética y contenido de carotenos totales en accesiones del germoplasma de yuca (Manihot esculenta Crantz). Acta Agron., 60(2), 97-107.
Dellaporta S.L., Wood J. & Hicks J.R., 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep., 1, 19-21.
Dice I.R., 1945. Measures of the amount of ecologic association between species. Ecology, 26, 297-302.
Ferguson M.E. et al., 2012. Molecular markers and their application to cassava breeding: past, present and future. Trop. Plant Biol., 5, 95-109.
Fregene M. et al., 1997. A molecular genetic map of cassava (Manihot esculenta Crantz). Theor. Appl. Genet., 95, 431-441.
Fregene M., Gutiérrez J.P. & Buitrago C., 2002. Protocols of cassava molecular genetic. CIAT Work Document. Cali,Colombia: CIAT.
Fregene M. et al., 2003. Simple Sequence Repeat marker diversity in cassava landraces: genetic diversity and differentiation in an asexually propagated crop. Theor. Appl. Genet., 107(7), 1083-1093.
Goudet J., 2001. FSTAT, Version. 1.2: A computer program to calculate F-statistics. Heredity, 86, 485-486.
Hurtado P. et al., 2008. Comparison of simple sequence repeats (SSR) and diversity array technology (DArT) markers for assessing genetic diversity in cassava. Plant Genet. Resour., 6(3), 208-214.
Kawuki R.S. et al., 2009. Identification, characterization and application of single nucleotide polymorphisms for diversity assessment in cassava (Manihot esculenta Crantz). Mol. Breed., 23(4), 669-684.
Kimura M. & Crow J.T., 1964. The number of alleles that can be maintained in a finite population. Genetics, 49, 725-738.
Kunkeaw S. et al., 2010. Genetic linkage map of cassava (Manihot esculenta Crantz) based on AFLP and SSR markers. Plant Breed., 129(1), 112-115.
Kunkeaw S. et al., 2011. Construction of a genetic linkage map using simple sequence repeat markers from expressed sequence tags for cassava (Manihot esculenta Crantz). Mol. Breed., 27(1), 67-75.
Lekha S.S., Teixeira da Silva J.A. & Pillai S.V., 2011. Genetic variability studies between released varieties of cassava and central Kerala cassava collections using SSR markers. J. Stored Prod. Postharvest Res., 2(4), 79-92.
Mba R.E.C. et al., 2001. Simple Sequence Repeats (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: towards an SSR-based molecular genetic map of cassava. Theor. Appl. Genet., 102, 21-31.
Milián M.D. et al., 2000. Caracterización, evaluación y conservación de la colección cubana de germoplasma de yuca (Manihot esculenta Crantz). In: Carvalho L.J.C.B., Thro A.M. & Vilarinhos A.D., eds. Proceeding of the 4th international scientific meeting cassava biotechnology network. Brasilia: Cassava Biotechnology Network.
MINAG, 2008. Instructivo Técnico sobre el Cultivo de la Yuca. La Habana: SEDGRI/AGRINFOR.
Minch E., 1996. MICROSAT. Version 1.4. Stanford, CA, USA: Stanford University Medical Centre.
Montero-Rojas M., Correa A.M. & Siritunga D., 2011. Molecular differentiation and diversity of cassava (Manihot esculenta) taken from 162 locations across Puerto Rico and assessed with microsatellite markers. AoB PLANTS, doi:10.1093/aobpla/plr010, (23/10/12).
Nei M., 1978. Estimation of average heterozygosity and genetics distances from a small number of individuals. Genetics, 89, 583-590.
Ojulong H., Labuschagne M.T., Herselman L. & Fregene M.A., 2010. Yield traits as selection indices in seedling populations of cassava. Crop Breed. Appl. Biotechnol., 10(3), 191-196.
Quenoille M.H., 1956. Notes on bias in estimation. Biometrika, 43, 353-360.
Raghu D. et al., 2007. Morphological and simple sequence repeats (SSR) based finger printing of South Indian cassava germplasm. Int. J. Integr. Biol., 2, 141-149.
Raji A.A. et al., 2009. Gene-based microsatellites for cassava (Manihot esculenta Crantz): prevalence, polymorphisms, and cross-taxa utility. BMC Plant Biol., 9, 118.
Rohlf F.J., 1998. NTYSYS-PC numerical taxonomy and multivariate analysis system. Version 2.02. New York, NY, USA: Applied Biostatistics Inc.
Roubus K., Moustakas M. & Aravanopoulos F.A., 2010. Molecular identification of Greek olive (Olea europaea) cultivars based on microsatellite loci. Genet. Mol. Res., 9, 1865-1876.
SAS Institute Inc., 1999. Statistic Analysis System. Version 8.0. Cary, NC, USA: SAS Institute Inc.
Siqueira M.V.B.M. et al., 2009. Genetic characterization of cassava (Manihot esculenta) landraces in Brazil assessed with simple sequence repeats. Genet. Mol. Biol., 32(1), 104-110.
Thotttapilly G., Crouch J.H. & Quin F.M., 1996. Overview of DNA marker research at IITA. DNA marker-assisted improvement of the staple crops of the sub-Saharan Africa. In: Jonathan H.C. & Abdou T., eds. Proceedings of the Workshop on DNA markers. Ibadan, Nigeria: IITA.
Turyagyenda L.F. et al., 2012. Genetic diversity among farmer-preferred cassava landraces in Uganda. Afr. Crop Sci. J., 20(S1), 15-30.
Valdés M.M., Albelo R.M. & Gallo G., 1991. La comunidad primitiva en Cuba. In: Ramos M.G. & Galán D.J., eds. Historia de Cuba. La Habana: Editorial Pueblo y Educación, 4-14.
Vázquez M., 2010. Alimentos esenciales en la cocina cubana: boniato, yuca y maíz, http://www.cubasolar.cu/biblioteca/energia/Energia43/HTML/Articulo11.htm, (21/04/2011).
Vásquez A. & López C., 2012. Identificación de polimorfismos en genes candidatos de resistencia en yuca (Manihot esculenta Crantz). Acta Agron., 61(2), 133-142.
Vekemans X. & Lefevre C., 1997. On the evolution of heavy metal tolerant populations in Armeria maritima: evidence from allozyme variation and reproductive barriers. J. Evol. Biol., 10, 175-199.
Vieira E.A. et al., 2011. Caracterização molecular de acessos de mandioca açucarados e não açucarados. Ciênc Agrotec, 35(3), 455-461.
Vimala B., Thushara R., Nambisan B. & Sreekumar J., 2011. Effect of processing on the retention of carotenoids in yellow-fleshed cassava (Manihot esculenta Crantz) roots. Int. J. Food Sci. Technol., 46, 166-169.
Weir B.S. & Cockerham C.C., 1984. Estimating F-statistics for the analysis of population structure. Evolution, 38, 1358-1370.
Whankaew S. et al., 2012. Characterization of microsatellite markers in cassava based on microsatellite-AFLP technique. Genet. Mol. Res., 11(2), 1319-1326.
Wright S., 1951. The genetical structure of populations. Ann. Eugenics, 15, 323-354.






