- Home
- Volume 15 (2011)
- numéro 2
- Beneficial effect of the rhizosphere microbial community for plant growth and health
View(s): 0 (0 ULiège)
Download(s): 0 (0 ULiège)
Beneficial effect of the rhizosphere microbial community for plant growth and health

Editor's Notes
Received on May 5, 2010; accepted on August 24, 2010
Résumé
Effet bénéfique de la communauté microbienne de la rhizosphère sur la croissance et la santé des plantes. La rhizosphère est le volume du sol situé au voisinage immédiat des racines des plantes et qui se caractérise par la présence d’exsudats racinaires (rhizodépôts). Ces exsudats sont utilisés par la microflore endémique en tant que signaux chimiques en plus d’être un substrat nutritif disponible pour la croissance et le développement de ces microorganismes dans la rhizosphère. Certaines de ces bactéries du sol, appelées PGPRs (Plant Growth Promoting Rhizobacteria), sont capables de coloniser les racines ou bien encore la rhizosphère, mais à la différence des autres bactéries rhizosphériques elles ont, en retour, un effet bénéfique sur la plante. Cet effet bénéfique peut être direct, ou indirect. La promotion directe de la croissance est le résultat du pouvoir d’acquisition des nutriments ou de la stimulation des hormones de la plante. D’autres mécanismes indirects, mais le plus souvent liés à la croissance des plantes, sont impliqués dans la réduction/suppression des pathogènes des plantes. Cet article décrit les différents mécanismes mis en jeu par les PGPRs dans leur environnement naturel pour influencer favorablement la croissance et la santé des plantes.
Abstract
Plant rhizosphere is the soil nearest to the plant root system where roots release large quantity of metabolites from living root hairs or fibrous root systems. These metabolites act as chemical signals for motile bacteria to move to the root surface but also represent the main nutrient sources available to support growth and persistence in the rhizosphere. Some of the microbes that inhabit this area are bacteria that are able to colonize very efficiently the roots or the rhizosphere soil of crop plants. These bacteria are referred to as plant growth promoting rhizobacteria (PGPR). They fulfil important functions for plant growth and health by various manners. Direct plant growth promotion may result either from improved nutrient acquisition and/or from hormonal stimulation. Diverse mechanisms are involved in the suppression of plant pathogens, which is often indirectly connected with plant growth. This paper describes the different mechanisms commonly used by most PGPR in their natural habitats to influence plant-growth and health.
Table of content
1. Introduction
1According to a general view, the rhizosphere includes plant roots and the surrounding soil. This is a wide and wise definition, already coined more than hundred years ago by Hiltner (1904). In that particular environment, very important and intensive interactions take place between the plant, soil, and microfauna. Biochemical interactions and exchanges of signal molecules between plants and soil microbes have been described and reviewed (Pinton et al., 2007). The rhizosphere inhabiting microorganisms compete for water, nutrients and space and sometimes improve their competitiveness by developing an intimate association with plant (Hartmann et al., 2009). These microorganisms play important roles in the growth and ecological fitness of their host. An understanding of the basic principles of rhizosphere microbial ecology, including the function and diversity of microorganisms that reside there, is necessary before soil microbial technology can be applied in the rhizosphere. Here we review different mechanisms commonly used by the beneficial rhizosphere bacteria to influence plant-growth and health in the natural environment.
2. The rhizosphere effect
2During seed germination and seedling growth, the developing plant interacts with a range of microorganisms present in the surrounding soil. As seeds germinate and roots grow through the soil, the release of organic material provides the driving force for the development of active microbial populations in a zone that includes plant root and surrounding soil in a few mm of thickness. This phenomenon is referred as the rhizosphere effect (Morgan et al., 2001).
3Broadly, there are three distinct components recognized in the rhizosphere; the rhizosphere per se (soil), the rhizoplane, and the root itself. The rhizosphere is thus the zone of soil influenced by roots through the release of substrates that affect microbial activity. The rhizoplane is the root surface, including the strongly adhering root particles. The root itself is a part of the system, because certain endophytic microorganisms are able to colonize inner root tissues (Bowen et al., 1999). The rhizosphere effect can thus be viewed as the creation of a dynamic environment where microbes can develop and interact.
3. Main characteristics of the root exudation process
4Root exudation is the release of organic compounds from living plant roots into the surrounding soil; it is an ubiquitous phenomenon (Jones et al., 1995). Roots release compounds via at least two potential mechanisms, and the rates of exudation sensu stricto vary widely among species and environmental conditions (Kochian et al., 2005). Exudates are transported across the cellular membrane and secreted into the surrounding rhizosphere. Plant products are also released from roots border cells and root border-like cells which separate from border as they grow (Bais et al., 2006). However, it is important to note that it is very difficult to identify root exudates with respect to the chemical composition and the concentration in the soil because of methodological difficulties (Stolp, 1988). At the moment of exudation and thereafter, the organic materials are subject to microbial attack, and thus cannot be enriched and separated from the roots in the natural environments. Data on the nature and quantity of root exudates have been obtained from sterile hydroponic cultures; but the results, however, are difficult to extrapolate to the natural conditions (Stolp, 1988). In this context, root exudation has been quantified by measuring the production of labelled CO2 in the rhizosphere of 14C-labelled plants, and it has been estimated that 12-40% of the total amount of carbohydrates produced by photosynthesis is released into the soil surrounding roots (Brimecombe et al., 2007). Root exudates are mainly composed of water soluble sugars, organic acids, and amino acids, but also contain hormones, vitamins, amino compounds, phenolics and sugar phosphate esters (Uren, 2001).
5Release of these low molecular weight compounds is a passive process along the steep concentration gradient which usually exists between the cytoplasm of intact root cells (millimolar range) and the external (soil) solution (micromolar range). Direct or passive diffusion through the lipid bilayer of the plasma membrane is determined by membrane permeability, which depends on the physiological state of the root cell and on the polarity of the compounds, facilitating the permeation of lipophilic exudates (Rudrappan et al., 2007). The efficiency of the exudation process may thus be enhanced by stress factors affecting membrane integrity such as nutrient deficiency, temperature extremes, or exudation stress (Ratnayale et al., 1978).
6It is assumed that both the qualitative and quantitative compositions of root exudates are affected by various environmental factors, including pH, soil type, oxygen status, light intensity, soil temperature, nutrient availability and the presence of microorganisms. These factors may have a greater impact on root exudation than differences due to the plant species (Singh et al., 2006).
7The proportion of carbon released from roots has been estimated to as much as 50% in the young plants (Whipps, 1990) but less in plants grown to maturity in the field (Jensen, 1993). The nature of exudates may also vary according to the growth stage of the plant. For instance, there are more carboxylates and root mucilage at the six leaf stage than earlier. On the other hand, nitrogen is also of considerable importance to nutrient cycling, usually as NH4+, NO3- (Wacquant et al., 1989), amino acids (Boulter et al., 1966), cell lysates, sloughed roots, and other root-derived debris. It is estimated at the maturity that the rhizodeposition of N amounted to 20% of the total plant nitrogen (Jensen, 1996). Root exudation is also largely dependent on the nutritional status of the plant regarding oligoelements. Low concentrations of some nutrients such as K+, Na+ and Mg++ readily stimulate the activity of major enzymes of the glycolytic pathway, namely phosphofructokinase and pyruvate kinase, which together regulate glycolysis in plant cells (Plaxton, 1996). Individual micronutrients are similarly important components of major enzymes, which regulate all biological processes in plants. It is clear from these considerations that low nutrient availability can constraint plant growth in many environments of the world, especially the tropics where soils are extremely deficient in these oligoelement nutrients (Pinton et al., 2007). Some species typically exude organic acid anions in response to P and Fe deficiency or phytosiderophores due to Fe and Zn deficiency (Haynes, 1990).
4. The rhizosphere-inhabiting microflora
4.1. Diversity
8The rhizosphere microflora include bacteria, fungi, nematodes, protozoa, algae and microarthrops (Raaijmakers et al., 2001). Of the soil microbes, 98% cannot be cultured. Their identification, characterization and the description of their role are therefore particularly difficult. Recently, nucleic acid based techniques including analysis of DNA and rRNA molecules from soil samples have revealed enormous diversity in the rhizosphere inhabiting microbial flora (Suzuki et al., 2006). The molecular methods used for soil microbial diversity are covered in the review by Nannipieri and collaborators (2003). The number of microbial species present in soil may vary from thousands to millions. Many studies indeed suggest that the Proteobacteria and the Actinobacteria form the most common of the dominant populations (>1%, usually much more) found in the rhizosphere of many different plant species (Singh et al., 2007). These groups contain many "cultured" members. They are the most studied of the rhizobacteria, and as such, contain the majority of the organisms investigated, both as beneficial microbial inoculants and as pathogens.
9The specific content of root exudates may create a niche that influences which microorganisms are to colonize the rhizosphere, thereby altering the composition and diversity of microorganisms colonizing the rhizosphere in a plant specific manner (Grayston et al., 1998). Plant species, plant developmental stage and soil type have thus been indicated as major factors determining the composition of rhizosphere microbial communities (Broeckling et al., 2008). That said, the extent to which the above-cited factors contribute to microbial communities is not fully understood and there are several contrasting reports in the literature indicating either plant or soil type as dominant factor (Nunan et al., 2005). Owing to the above statement, it can be generalized that the diversity and predominance of rhizosphere microbial population depend on a number of abiotic and biotic factors prevailing in that particular ecological niche (Figure 1).
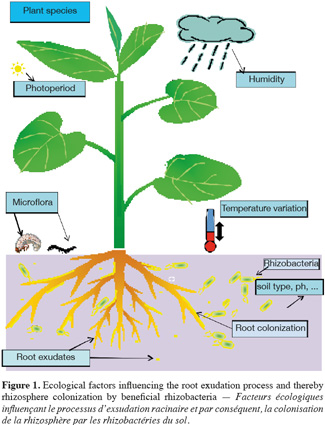
4.2. Population level
10Studies based on the use of growth media steadily showed that bacterial populations residing in the rhizosphere are several orders of magnitude larger than those residing in bulk soils. Rhizosphere bacteria concentration can reach between 1010 and 1012 cells per gram of soil (Foster, 1988), and they are transferred to various associated environments including plants, foods, animals, marine and freshwater habitats (Buée et al., 2009). Only few groups of these bacteria are considered to be soilborne, probably because non-spore forming bacteria cannot survive well in soil for long periods.
11The effect of root exudates depends on the distance that they can diffuse away from rhizoplane (Gupta et al., 2002). Bacterial communities are not uniformly distributed along root axes, and differ between root zones. Distinct bacterial community compositions are obtained by molecular fingerprints in different root zones, like those of emerging roots and root tips, elongating roots, sites of emergence of lateral roots, and older roots (Yang et al., 2000). It has been proposed that populations residing in the rhizosphere oscillate along root axes in a wave-like fashion (Semenov et al., 1999). Accordingly, bacterial communities temporarily profit from the nutrients released by younger roots in the root hair zones, and wave-like fluctuations in bacterial cell numbers can be explained by death and lysis of bacterial cells upon starvation when nutrients become depleted, followed by cell divisions in surviving and thus viable populations as promoted by the release of nutrients from dead and decaying cells (Semenov et al., 1999). Bacterial communities in rhizosphere soils are thus not static, but will fluctuate over time in different root zones.
4.3. The rhizosphere as a battle field
12The number and diversity of microorganisms are related to the quantity and quality of the exudates but also to the outcome of the microbial interactions that occur in the rhizosphere (Somers et al., 2004). Soil biota (bacteria, fungi, micro-fauna and the plant root) are themselves embedded in food webs and thus interactions with consumers or predators in the microbial as well as macro- and mesofaunal world are important to understand rhizosphere processes. A high number of soil microbes attained properties enabling them to interact more efficiently with roots and withstand the quite challenging conditions of rhizosphere life. The rhizosphere inhabiting microorganisms compete each other for water, nutrients and space and sometimes improve their competitiveness by developing an intimate association with plant. This process can be regarded as an ongoing process of micro-evolution in low-nutrient environments, which are quite common in natural ecosystems (Schloter et al., 2000).
5. Plant-bacteria interactions in the rhizosphere
13Microorganisms present in the rhizosphere play important roles in ecological fitness of their plant host. Important microbial processes that are expected to occur in the rhizosphere include pathogenesis and its counterpart, plant protection/growth promotion, as well as the production of antibiotics, geochemical cycling of minerals and plant colonization (Kent et al., 2002). Plant-microbe interactions may thus be considered beneficial, neutral, or harmful to the plant, depending on the specific microorganisms and plants involved and on the prevailing environmental conditions (Bais et al., 2006). Exploring these microorganisms by unravelling their possible relationships with plants has launched a new and fascinating area of investigations in the rhizosphere research.
5.1. Pathogenic interactions
14Roots exudates can attract beneficial organisms (see below), but they can also be equally attractive to pathogenic populations (Schroth et al., 1964), that many express virulence on only a limited number of host species. Many pathogenic organisms, bacteria as well as fungi, have coevolved with plants and show a high degree of host specificity (Raaijmakers et al., 2009). In nature however, plant disease is the exception rather than the rule because the conditions that are optimized for the plant growth may not be favourable for pathogens (Paulitz et al., 2001).
15Plants are not defenceless. In fact, it is estimated that only about 2% of the known fungal species are able to colonize plants and cause disease (Buchanan et al., 2000). Even though plants are in permanent contact with potential pathogens such as fungi, bacteria or viruses, successful infection is rarely established. Such a general resistance against most pathogens has been named “horizontal resistance” or “non-host-resistance” (Heath, 1981). This reflects the fact that the plant is not a suitable target for infection by a specific pathogen due to preformed, passive resistance mechanisms resulting in “basic incompatibility”. These resistance mechanisms comprise structural barriers and toxic compounds that are present in the unaffected, healthy plant and limit successful infection to specialized pathogens that have the ability to overcome these factors and therefore exhibit “basic compatibility”. If contact is nevertheless established with the plant tissue, pathogens are often confronted with preformed chemical components named phytoanticipins (van Etten et al., 1994). This term comprises a variety of compounds produced by different biosynthetic pathways which possess antimicrobial properties. These low molecular weight secondary metabolites are mainly stored in inactive form in the vacuoles or organelles and are released upon destruction of the cells. Since destroying the integrity of the plant tissue is part of the colonization strategy by fungi, phytoanticipins represent an important resistance mechanism against these pathogens.
16However, in some instances, pathogens can overcome the pre-formed barriers and develop virulent infection processes leading to plant disease. Plant diseases play a direct role in the destruction of natural resources in agriculture. In particular, soil-borne pathogens cause important losses, fungi being the most aggressive. The extent of their harmful effects ranges from mild symptoms to catastrophes where large fields planted with agricultural crops are destroyed. Thus, they are major and chronic threats to food production and ecosystem stability worldwide. Common and well investigated bacterial agents include Gram- bacteria Erwinia carotovora, Pseudomonas, Ralstonia spp. and the Gram+ bacterium Streptomyces scabies. The fungal and oomycete phytopathogens include members of Fusarium, Phytophthora, Pythium, Rhizopus, Rhizoctonia and Verticillium (Tournas et al., 2005). From the forest pathogens, among the most important are the filamentous fungi Heterobasidion and Armillariella (Asiegbu et al., 2005), and Phytophthora spp. (Rizzo et al., 2005).
5.2. Beneficial microorganisms and modes of action
17Plant-beneficial microbial interactions can be roughly divided into three categories. First, those microorganisms that, in association with plants, are responsible for its nutrition (i.e., microorganisms that can increase the supply of mineral nutrients to the plant). In this case, while most may not directly interact with the plant, their effects on soil biotic and abiotic parameters certainly have an impact on plant growth. Second, there is a group of microorganisms that stimulate plant growth indirectly by preventing the growth or activity of pathogens. Such microorganisms are referred to as biocontrol agents, and they have been well documented. A third group involves those microorganisms responsible for direct growth promotion, for example, by production of phytohormones. There has been a large body of literature describing potential uses of plant associated bacteria as agents stimulating plant growth and managing soil and plant fitness (Welbaum et al., 2004). On another hand, apparently neutral interactions are found extensively in the rhizosphere of all crop plants. Saprophytic microorganisms are responsible for many vital soil processes, such as decomposition of organic residues in soil and associated soil nutrient mineralization or turnover processes. Whereas these organisms do not appear to benefit or harm the plant directly (hence the term neutral), their presence is obviously vital for soil dynamic, and their absence would clearly influence plant health and productivity (Brimecombe et al., 2007).
18Rhizosphere-living bacteria that exert a global beneficial effect on plant growth are referred as plant growth promoting rhizobacteria (PGPR) (Kloepper et al., 1978). The number of bacterial species identified as PGPR increased recently as a result of the numerous studies covering a wider range of plant species and because of the advances made in bacterial taxonomy and the progress in our understanding of the different mechanisms of action of PGPR. Presently, PGPR include representatives from very diverse bacterial taxa (Lucy et al., 2004) and in the following sections we are not giving a thorough description of all the genera and species of PGPR, but rather a few examples to illustrate the diversity and modes of action of these beneficial bacteria. Diverse PGPR strains have been used successfully for crop inoculations These comprise members of the bacterial genera Azospirillum (Cassán et al., 2008), Bacillus (Jacobsen et al., 2004), Pseudomonas (Loper et al., 2007), Rhizobium (Long, 2001), Serratia (De Vleeschauwer et al., 2007), Stenotrophomonas (Ryan et al., 2009), and Streptomyces (Schrey et al., 2008). Some fungi belonging to the genera Ampelomyces, Coniothyrium, and Trichoderma have also been described to be beneficial for the host plant (Harman et al., 2004). The modes of action of PGPR involve complex mechanisms to promote plant growth, development and protection. Important among them are biofertilization (increasing the availability of nutrients to plant), phytostimulation (plant growth promoting, usually by the production of phytohormones) and biocontrol (controlling diseases, mainly by the production of antibiotics and antifungal metabolites, lytic enzymes and induction of plant defense responses). Pseudomonas and Bacillus genera are the most commonly investigated PGPR, and often the dominating bacterial groups in the rhizosphere (Morgan et al., 2005). One has to mention that, in many cases of individual beneficial plant-microbe interactions, several mechanisms are involved (Müller et al., 2009). Ad planta, direct mechanisms of plant growth promotion are difficult to differentiate from disease suppression and the relative importance on a specific mechanism can vary within different pathosystems (Chet et al., 2002).
19Colonization. In all successful plant-microbe interactions, the competence to colonize plant habitats is important (Lugtenberg et al., 2002; Kamilova et al., 2005). Single bacterial cells can attach to surfaces and, after cell division and proliferation, form dense aggregates commonly referred to as macrocolonies or biofilms. Steps of colonization include attraction, recognition, adherence, invasion (only endophytes and pathogens), colonization and growth, and several strategies to establish interactions. Plant roots initiate crosstalk with soil microbes by producing signals that are recognized by the microbes, which in turn produce signals that initiate colonization (Berg, 2009). PGPR reach root surfaces by active motility facilitated by flagella and are guided by chemotactic responses (Pinton et al., 2007). This implies that PGPR competence highly depends either on their abilities to take advantage of a specific environment or on their abilities to adapt to changing conditions or plant species. As an example, when strain S499 of Bacillus subtilis was applied to plant seedlings, it showed more distinct but effective colonization of the root system of two distinct plants (Figure 2). In most cases, the population of many PGPR inoculants actually declines progressively in time after inoculation from 107-109 cells per gram dry soil to 105-106 cells per gram dry soil after 2-3 weeks (DeFlaun et al., 1993). Nevertheless this population threshold is often sufficient to provide beneficial effects (Raaijmakers et al., 2002). Rhizosphere competence of biocontrol agents thus involves effective root colonization combined with the ability to survive and proliferate along growing plant roots over a large time period, in the presence of the indigenous microflora (Weller, 1988; Lugtenberg et al., 1999).
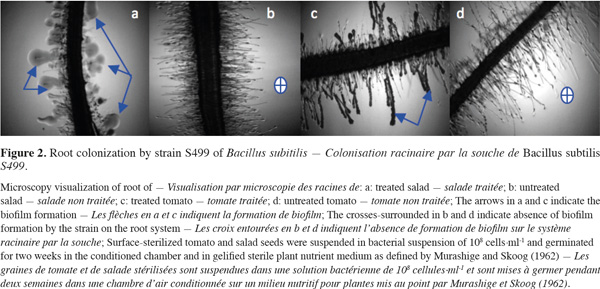
20Pathogen inhibition. Bacteria and fungi live around roots and feed on root exudates and dead root cells. Competition between microbial species in this area is stiff. In the battle for establishment and persistence in the niche, bacteria use several strategies.
21Antagonism. Root colonization not only results in high PGPR population densities on the root system, it also functions as the delivery system of antagonistic metabolites that are involved in direct inhibition of plant pathogens (Shoda, 2000; Raaijmakers et al., 2002). It includes antibiosis i.e. the inhibition of microbial growth by diffusible antibiotics and volatile organic compounds, toxins, and biosurfactants, and parasitism that may involve production of extracellular cell wall-degrading enzymes such as chitinases and β-1,3-glucanase (Compant et al., 2005; Haas et al., 2005). The degradation of pathogenicity factors of the pathogen such as toxins by the beneficial organism has also been reported as protective mechanism (Haas et al., 2005). To demonstrate the role of antibiotics in biocontrol, mutants impaired in biosynthesis or over-producing mutants have been used together with, in some cases, the use of reporter genes or probes to show efficient production of the compound in the rhizosphere. As example, Bacillus subtilis strains produce a variety of powerful antifungal metabolites, e.g., zwittermicin-A, kanosamine and lipopeptides from the surfactin, iturin and fengycin families (Emmert et al., 1999; Ongena et al., 2006). Dunne and collaborators (2000) showed that overproduction of extracellular protease in the mutant strains of Stenotrophomonas maltophilia W81 resulted in improved biocontrol of Pythium ultimum. Excretion of chitinases and glucanases by species of Trichoderma and Streptomyces has also been shown to play an important role in mycoparasitism of phytopathogenic fungi (Whipps, 2001).
22Competition. Competition for resources such as nutrients and oxygen occurs generally in soil between soil-inhabiting organisms. For biocontrol purpose, it occurs when the antagonist directly competes with pathogens for these resources. Root inhabiting microorganisms compete for suitable sites at the root surfaces. Competition for nutrients, especially for carbon, is assumed to be responsible for the well-known phenomenon of fungistasis characterizing the inhibition of fungal spore germination in soil (Alabouvette et al., 2006). Given the relative abundance of substrates in the rhizosphere, the efficiency of nutrient uptake and catabolism by bacteria is a key factor in competitiveness (Chin-A-Woeng et al., 2003). The capacity for rapid growth when substrates are encountered is not the only factor affecting rhizosphere competence, as rhizobacteria deploy many other metabolic strategies. For example, the capacity for extracellular conversion of glucose to gluconic acid and 2-ketogluconic acid enables some bacteria, including several species of Pseudomonas to sequester glucose effectively and gives a competitive advantage over microorganisms that lack the ability to utilise these compounds (Gottschalk, 1986).
23Competition for trace elements, such as iron, copper, zinc, manganese, etc. also occurs in soils. For example, iron is an essential growth element for all living organisms and the scarcity of its bio-available form in soil habitats results in a furious competition (Loper et al., 1997). Siderophores, low molecular weight compounds with high iron affinity, are produced by some microorganisms (also by most biocontrol agents) to solubilize and competitively acquire ferric ion under iron-limiting conditions, thereby making iron unavailable to other soil microorganisms which cannot grow for lack of it (Loper et al., 1997; Haas et al., 2005).The bacterium that originally synthesized the siderophores takes up the iron siderophore complex by using a receptor that is specific to the complex and is located in the outer cell membrane. Suppression of soil borne plant pathogens by siderophore producing Pseudomonads has been reported in some instances (Loper, 1988; Weger et al., 1988; Buysens et al., 1996).
24Induced resistance. Plant-associated bacteria can reduce the activity of pathogenic microorganisms not only through microbial antagonisms, but also by activating the plant to better defend itself, a phenomenon termed “induced systemic resistance”, “ISR” (Shoda, 2000; Van Loon, 2007). Sometimes, the mechanism of ISR elicited by PGPR overlaps partly with that of pathogen-induced systemic acquired resistance (SAR). Both ISR and SAR represent a state of enhanced basal resistance of the plant that depends on signalling compounds such as jasmonic acid, ethylene and salicylic acid (Van Loon, 2007). Expression of natural defense reaction against stresses from biotic or abiotic origin is exhibited by all plants, such as:
25- physical stresses (heat or frost);
26- inoculation by pathogenic or non-pathogenic organisms;
27- chemical molecules from natural or synthetic origins (Alabouvette et al., 2006).
28Early recognition of the aggressor by the plant is one of the mechanisms involved in elicitation of plant defense reactions (Lugtenberg et al., 2002). Recognition of the aggressor immediately initiates a cascade of molecular signals and the transcription of many genes, which eventually results in the production of defence molecules by the host plant (van Loon, 2000). Such defence molecules include phytoalexins, pathogenesis-related (PR) proteins (such as chitinases, β-1,3-glucanases, proteinase inhibitors, etc.) and lignin for reinforcement of cell walls (van Loon, 2000). In fact, cell wall thickenings, wall appositions or rapid death of the injured plant cells resulting in necrosis of the immediate adjacent tissues are barriers which cut the pathogen off its nutrients and contribute to slowing down of the fungus progressive invasion (Lugtenberg et al., 2002; Alabouvette et al., 2006).
29Plant growth promotion
30Phytostimulation. Phytostimulation enhances plant growth in a direct way. In the processes of plant growth, phytohormones [e.g., production of indole-3-acetic acid (IAA), auxins, cytokinins, and gibberellins] play an important role. These hormones can be synthesized by the plant themselves but also by their associated microorganisms such as Azospirillum spp., besides having nitrogen-fixing ability (Steenhoudt et al., 2000). Species of Pseudomonas and Bacillus can produce as yet not well characterized phytohormones or growth regulators that cause crops to have greater amounts of fine roots which have the effect of increasing the absorptive surface of plant roots for uptake of water and nutrients. The phytohormones they produce include indole-acetic acid, cytokinins, gibberellins and inhibitors of ethylene production. Indole-3-acetic acid is a phytohormone which is known to be involved in root initiation, cell division, and cell enlargement (Salisbury, 1994). This hormone is very commonly produced by PGPRs (Barazani et al., 2001). Auxins are quantitatively the most abundant phytohormones secreted by Azospirillum, and it is generally agreed that their production, rather than nitrogen-fixation, is the major factor responsible for the stimulation of rooting and, hence, enhanced plant growth (Bloemberg et al., 2001). Furthermore, plant-associated bacteria can influence the hormonal balance of the plant. Ethylene is an important example to show that the balance is most important for the effect of hormones: at low levels, it can promote plant growth in several plant species including Arabidopsis thaliana, while it is normally considered as an inhibitor of plant growth and known as a senescence hormone (Pierik et al., 2006).
31The general effect on the plant can be direct, that is through plant growth promotion, or indirect, that is through improving plant nutrition via the better development of the roots, and it is difficult to distinguish between them. The elevation of root IAA level in lodgepole pine plantlets, inoculated with Paenibacillus polymyxa, and, of dihydroxyzeatin riboside root concentration in plants inoculated with Pseudomonas fluorescens (Fuentes-Ramirez et al., 2005), might be attributed to the induction of plant hormone synthesis by the bacteria. However, the uptake of bacterial synthesized phytohormones can not be excluded, since both P. polymyxa and Pseudomonas produce IAA and cytokinins in vitro (Fuentes-Ramirez et al., 2005).
32Biofertilization. The mechanisms by which PGPR increases crop performance is not well understood. There are several PGPR inoculants currently commercialized that seem to promote growth through at least one mechanism; suppression of plant disease (termed bioprotectants), phytohormone production (termed biostimulants), or improved nutrient acquisition (termed biofertilizers). The mode of action of PGPR by biofertilizers act either, directly by helping to provide nutrient to the host plant, or indirectly by positively influencing root growth and morphology or by aiding other beneficial symbiotic relationships (Vessey, 2003). The most prominent example is bacterial nitrogen fixation. The symbiosis between rhizobia and its legume host plants is an important example for plant growth-promoting rhizobacteria (PGPR). Bacteria of this group metabolize root exudates (carbohydrates) and in turn provide nitrogen to the plant for amino acid synthesis. The ability to fix nitrogen also occurs in free-living bacteria like Azospirillum, Burkholderia, and Stenotrophomonas (Dobbelare et al., 2003). Biofertilization accounts for approximately 65% of the nitrogen supply to crops worldwide (Bloemberg et al., 2001). Another nutrient is sulfate, which can be provided to the plant via oxidation by bacteria (Banerjee et al., 2002). Bacteria may contribute to plant nutrition by liberating phosphorous from organic compounds such as phytates and thus indirectly promote plant growth (Unno et al., 2005). Azospirillum treatment resulted in enhancement of root growth and activities (e.g., acidification of the root surroundings) that increases phosphorous and other macroelements and microelements uptake (Dobbelaere et al., 2007). Recently, De Werra and collaborators (2009) showed that the ability of Pseudomonas fluorescens CHA0 to acidify its environment and to solubilize mineral phosphate is strongly dependent on its ability to produce gluconic acid.
6. Conclusion
33The rhizosphere is the zone of soil surrounding a plant root where the biology and chemistry of the soil are influenced by the root. As plant roots grow through soil they mostly release water soluble compounds such as amino acids, sugars and organic acids that supply food for the microorganisms. High levels of exudates in the rhizosphere attract a plethora of microorganisms to a larger extend than elsewhere in the soil. The composition and pattern of root exudates affect microbial activity and population numbers. Plant species, plant developmental stage and soil type have been indicated as major factors determining the composition of rhizosphere microbial communities (Broeckling et al., 2008). As shown in many studies, there is no general decision about the key player: the diversity and predominance of rhizosphere microbial population depend on a number of abiotic and biotic factors of a particular ecological niche.
34A better understanding of the basic principles of the rhizosphere ecology, including the function and diversity of inhabiting microorganisms is on the way but further knowledge is necessary to optimize soil microbial technology to the benefit of plant-growth and health in the natural environment. In sum, this can constitute overwhelming evidence indicating that an ever exploitation of plant growth promoting rhizobateria (PGPR) can be a true success story in sustainable agriculture. As a consequence, current production methods in agriculture, e.g., the improper use of chemical pesticides and fertilizers creating a long list of environmental and health problems, will reduce.
35Acknowledgements
36Venant Nihorimbere is recipient of a grant from the Belgian Technical Cooperation (BTC/CTB). Marc Ongena is Research Associate at the F.R.S.-FNRS (National Funds for Scientific Research, Belgium).
Bibliographie
Alabouvette C., Olivain C. & Steinberg C., 2006. Biological control of plant diseases: the European situation. Eur. J. Plant Pathol., 114, 329-341.
Asiegbu F.O. & Nahalkova J.L.G., 2005. Pathogen-inducible cDNAs from the interaction of the root rot fungus Heterobasidion annosum with Scots pine (Pinus sylvestris L.). Plant Sci., 168, 365-372.
Bais H.P. et al., 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Ann. Rev. Plant Biol., 57, 233-266.
Banerjee M. & Yesmin L., 2002. Sulfur-oxidizing plant growth promoting Rhizobacteria for enhanced canola performance. US Patent 20080070784.
Barazani O. & Friedman J., 2001. Allelopathic bacteria and their impact on higher plants. Crit. Rev. Microbiol., 27, 41-55.
Berg G., 2009. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol., 84, 11-18.
Bloemberg G.V. & Lugtenberg B.J.J., 2001. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol., 4, 343-350.
Boulter D., Jeremy J.J. & Wilding M., 1966. Amino acids liberated into the culture medium by pea seedling roots. Plant Soil, 24, 121-127.
Bowen G. & Rovira A., 1999. The rhizosphere and its management to improve plant growth. Adv. Agron., 66, 1-102.
Brimecombe M.J., De Leij F.A.A.M. & Lynch J.M., 2007. Rhizodeposition and microbial populations. In: Pinton R., Veranini Z. & Nannipieri P., eds. The rhizosphere biochemistry and organic substances at the soil-plant interface. New York, USA: Taylor & Francis Group.
Broeckling C.D. et al., 2008. Root exudates regulate soil fungal community composition and diversity. Appl. Environ. Microbiol., 74, 738-744.
Buchanan R., Gruissem W. & Jones R.L., 2000. Biochemistry and molecular biology of plants. Rockville, MD, USA: American Society of Plant Biologists.
Buée M. et al., 2009. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil, 321, 189-212.
Buysens S., Heungens K., Poppe J. & Höfte M., 1996. Involvement of pyochelin and pyoverdin in suppression of Pythium-induced damping-off of tomato by Pseudomonas aeruginosa 7NSK2. Appl. Environ. Microbiol., 62, 865-871.
Cassán F. & García Salamone I., 2008. Azospirillum sp.: cell physiology, plant response, agronomic and environmental research in Argentina. BuenosAires: Asociacion Argentina de Microbiologia.
Chet I. & Chernin L., 2002. Biocontrol, microbial agents in soil. In: Bitton G., ed. Encyclopedia of environmental microbiology. New York, USA: Willey, 450-465.
Chin-A-Woeng T.F.C., Bloemberg G.V. & Lugtenberg B.J., 2003. Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol., 157, 503-523.
Compant S. et al., 2005. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol., 71, 4951-4959.
De Vleeschauwer D. & Höfte M., 2007. Using Serratia plymuthica to control fungal pathogens of plants. CAB Rev., 2, 46.
De Werra P., Péchy-Tarr M., Keel C. & Maurhofer M., 2009. Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol., 75, 4162-4174.
DeFlaun M.F. & Gerba C.P., 1993. Monitoring recombinant DNA microorganisms and viruses in soil. In: Metting F.B.J., eds. Soil microbial ecology: application in agricultural and environmental management. Washington: Marcel Dekker Inc., 131-150.
Dobbelare S., Vanderleyden J. & Okon Y., 2003. Plant-growth promoting effects of diazotrophs in the rhizosphere. Crit. Rev. Plant Sci., 22, 107-149.
Dobbelaere S. & Okon Y., 2007. The plant growth promoting effects and plant responses. In: Elmerich C. & Newton W.E., eds. Associative and endophytic nitrogen-fixing bacteria and cyanobacterial associations (Nitrogen fixation: origins, applications and research progress). Heidelberg, Germany: Springer, 145-170.
Dunne C., Moenne-Loccoz Y., de Bruijn F.J. & O’Gara F., 2000. Overproduction of an inducible extracellular serine protease improves biological control of Pythium ultimum by Stenotrophomonas maltophilia strain W81. Microbiology, 146, 2069-2078.
Emmert E.A.B. & Handelsman J., 1999. Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol. Lett., 171, 1-9.
Foster R.C., 1988. Microenvironments of soil microorganisms. Biol. Fertil. Soils, 6, 189-203.
Fuentes-Ramirez L.E. & Caballero-Mellado J., 2005. Bacterial biofertilizers. In: Siddiqui Z.A., ed. PGPR: biocontrol and biofertilization. Dordrecht, The Netherlands: Springer, 143-172.
Gottschalk G., 1986. Bacterial metabolism. Berlin; Heidelberg, Germany; New York, USA: Springer.
Grayston S.J., Wang S.Q., Campbell C.D. & Edwards A.C., 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem., 30, 369-378.
Gupta R. & Mukerji K.G., 2002. Root exudate-biology. In: Mukerji K.G., Manoharachary C., Chamola B.P., eds. Techniques in mycorrhizal studies. Dordrecht, The Netherlands: Kluwer Academic Publishers, 103-131.
Haas D. & Défago G., 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol., 3, 307-319.
Harman G.E. et al., 2004. Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol., 2, 43-56.
Hartmann A., Schmid M., van Tuinen D. & Berg G., 2009. Plant-driven selection of microbes. Plant Soil, 321, 235-257.
Haynes R.J., 1990. Active ion uptake and maintenance of cationanion balance: a critical examination of their role in regulating rhizosphere pH. Plant Soil, 126, 247-264.
Heath M.C., 1981. A generalized concept of host-parasite specificity. Phytopathology, 71, 1121-1123.
Hiltner L., 1904. Über neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter besonderer Berücksichtigung der Gründüngung und Brache. Arb. Dtsch. Landwirtsch. Ges., 98, 59-78.
Jacobsen B.J., Zidack N.K. & Larson B.J., 2004. The role of Bacillus-based biological control agents in integrated pest management systems: plant diseases. Phytopathology, 94, 1272-1275.
Jensen B., 1993. Rhizodeposition by 14CO2-pulse-labelled spring barley grown in small field plots on sandy loam. Soil Biol. Biochem., 25, 1553-1559.
Jensen E.S., 1996. Rhizodeposition of N by pea and barley and its effects on soil N dynamics. Soil Biol. Biochem., 28, 65-71.
Jones D.L. & Darrah P.R., 1995. Influx and efflux of organic acids across the soil-root interface of Zea mays L. and its implications in rhizosphere C flow. Plant Soil, 173, 103-109.
Kamilova F. et al., 2005. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol., 7, 1809-1817.
Kent A.D. & Triplett E.W., 2002. Microbial communities and their interactions in soil and rhizosphere ecosystems. Ann. Rev. Microbiol., 56, 211-236.
Kloepper J.W. & Schroth M.N., 1978. Plant growth promoting rhizobacteria on radish. In: Station de pathologie végétale et phyto-bacteriologie, ed. Proceedings of the 4th Conference plant pathogenic bacteria, Angers, INRA, 879-882.
Kochian L., Piñeros M. & Hoekenga O., 2005. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil, 274, 175-195.
Long S.R., 2001. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol., 125, 69-72.
Loper J.E., 1988. Role of fluorescent siderophore production in biological control of Pythium ultirnum by a Pseudomonas fluorescens strain. Phytopathology, 78, 166-172.
Loper J.E. & Henkels M.D., 1997. Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl. Environ. Microbiol., 63, 99-105.
Loper J.E. & Gross H., 2007. Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf-5. Eur. J. Plant Pathol., 119, 265-278.
Lucy M., Reed E. & Glick B.R., 2004. Applications of free living plant growth promoting rhizobacteria. Antonie Van Leeuwenhoek, 86, 1-25.
Lugtenberg B.J.J. & Dekkers L.C., 1999. What makes Pseudomonas bacteria rhizosphere competent. Environ. Microbiol., 1, 9-13.
Lugtenberg B.J.J., Chin-A-Woeng T.F.C. & Bloemberg G.V., 2002. Microbe-plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek, 81, 373-383.
Morgan J.A.W. & Whipps J.M., 2001. Methodological approaches to the study of rhizosphere carbon flow and microbial population dynamics. In: Pinton A., Varanini Z. & Nannipieri P., eds. The rhizosphere. Biochemistry and organic substances at the soil-plant interface. New York, USA: Marcel Dekker, 373-409.
Morgan J.A., Bending G.D. & White P.J., 2005. Biological costs and benefits to plant-microbe interactions in the rhizosphere. J. Exp. Bot., 56, 1729-1739.
Müller H. et al., 2009. Quorum-sensing effects in the antagonistic rhizosphere bacterium Serratia plymuthica HRO-C48. FEMS Microbiol. Ecol., 67, 468-478.
Murashige T. & Skoog F., 1962. A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant, 15, 473-497.
Nannipieri P. et al., 2003. Microbial diversity and soil functions. Eur. J. Soil Sci., 54, 655-670.
Nunan N. et al., 2005. Links between plant and rhizoplane bacterial communities in grassland soils, characterized using molecular techniques. Appl. Environ. Microbiol., 71, 6784-6792.
Ongena M. & Thonart P., 2006. Resistance induced in plants by non-pathogenic microorganisms: elicitation and defense responses. In: Teixeira da Silva J.A., ed. Floriculture, ornamental and plant biotechnology: advances and topical issues. London: Global Science Books, 447-463.
Paulitz T.C. & Bélanger R.R., 2001. Biological control in greenhouse systems. Annu. Rev. Microbiol., 39, 103-133.
Pierik R. et al., 2006. The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci., 11, 176-183.
Pinton R., Veranini Z. & Nannipieri P., 2007. The rhizosphere. Biochemistry and organic substances at the soil-plant interface. New York, USA: Taylor & Francis Group, LLC.
Plaxton W.C., 1996. The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol., 47, 185-214.
Raaijmakers J.M. & Weller D.M., 2001. Exploiting genotypic diversity of 2,4-diacetylphloroglucinol-producing Pseudomonas spp.: characterization of superior root-colonizing P. fluorescens strain Q8r1-96. Appl. Environ. Microbiol., 67, 2545-2554.
Raaijmakers J.M., Vlami M. & de Souza J.T., 2002. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek, 81, 537-547.
Raaijmakers J.M. et al., 2009. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil, 321, 341-361.
Ratnayale M., Leonard R.T. & Menge A., 1978. Root exudation in relation to supply of phosphorus and its possible relevance to mycorrhizal infection. New Phytol., 81, 543-552.
Rizzo D.M., Garbelotto M. & Hansen E.A., 2005. Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol., 43, 309-335.
Rudrappan T., Quinn W.J., Stanley-Wall N.R. & Bais H.P., 2007. A degradation product of the salicylic acid pathway triggers oxidative stress resulting in down-regulation of Bacillus subtilis biofilm formation on Arabidopsis thaliana roots. Planta, 226, 283-297.
Ryan R.P. et al., 2009. Versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Microbiol. Rev., 7, 514-525.
Salisbury F.B., 1994. The role of plant hormones. In: Wilkinson R.E., ed. Plant-environment interaction. New York, USA: Dekker, 39-81.
Schloter M., Lebuhn M., Heulin T. & Hartmann A., 2000. Ecology and evolution of bacterial microdiversity. FEMS Microbiol. Rev., 24, 647-660.
Schrey S.D. & Tarkka M.T., 2008. Friends and foes: streptomycetes as modulators of plant disease and symbiosis. Antonie Van Leeuwenhoek, 94, 11-19.
Schroth M.N. & Hildebrand D.C., 1964. Influence of plant exudates on root-infecting fungi. Annu. Rev. Phytopathol., 2, 101-132.
Semenov A.M., van Bruggen A.H.C. & Zelenev V.V., 1999. Moving waves of bacterial populations and total organic carbon along roots of wheat. Microbiol. Ecol., 37, 116-128.
Shoda M., 2000. Bacterial control of plant diseases. J. Biosci. Bioeng., 89, 515-521.
Singh G. & Mukerji K.G., 2006. Root exudates as determinant of rhizospheric microbial biodiversity. In: Mukerji K.G., Manoharachary C. & Singh J., eds. Microbial activity in the rhizosphere. Berlin- Heidelberg, Germany: Springer-Verlag, 39-53.
Singh S. et al., 2007. Evaluation of mulching, intercropping with Sesbania and herbicide use for weed management in dry-seeded rice (Oryza sativa L.). Crop Prot., 26, 518-524.
Somers E., Vanderleyden J. & Srinivasan M., 2004. Rhizosphere bacterial signalling: a love parade beneath our feet. Crit. Rev. Microbiol., 30, 205-235.
Steenhoudt O. & Vanderleyden J., 2000. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol. Rev., 24, 487-506.
Stolp H., 1988. Microbial ecology: organisms, habitats, activities. New York, USA: Cambridge University Press.
Suzuki M. et al., 2006. Biosynthesis and secretion of mugineic acid family phytosiderophores in zinc-deficient barley. Plant J., 48, 85-97.
Tournas V.H. & Katsoudas E., 2005. Mould and yeast flora in fresh berries, grapes and citrus fruits. Int. J. Food Microbiol., 105, 11-17.
Unno Y. et al., 2005. Plant growth promotion abilities and microscale bacterial dynamics in the rhizosphere of lupin analysed by phytate utilization ability. Environ. Microbiol., 7, 396-404.
Uren N.C., 2001. Types, amounts and possible functions of compounds released into the rhizosphere by soil-grown plants. In: Pinton R., Varanini Z. & Nannipieri P., eds. The rhizosphere. Biochemistry and organic substances at the soil-plant interface. New York, USA: Marcel Dekker, 19-40.
van Etten H.D., Mansfield J.W., Bailey J.A. & Farmer E.E., 1994. Two classes of plant antibiotics: phytoalexins versus phytoanticipins. Plant Cell, 6, 1191-1192.
van Loon J.C., 2000. Induced resistance. In: Slusarenko A.J., Fraser R.S.S. & Van Loon J.C., eds. Mechanisms of resistance to plant diseases. Dordrecht, The Netherlands: Kluwer Academic Publishers, 521-574.
Van Loon L.C., 2007. Plant responses to plant growth promoting bacteria. Eur. J. Plant Pathol., 119, 243-254.
Vessey J.K., 2003. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil, 255, 571-586.
Wacquant J.P., Ouknider M. & Jacquard P., 1989. Evidence for a periodic excretion of nitrogen by roots of grass-legume associations. Plant Soil, 116, 57-68.
Weger L.A. et al., 1988. Siderophore-mediated uptake of Fe3+ by the plant growth-stimulating Pseudomonas putida strain WCS358 and by other rhizosphere microorganisms. J. Bacteriol., 170, 4693-4698.
Welbaum G., Sturz A.V., Dong Z. & Nowak J., 2004. Fertilizing soil microorganisms to improve productivity of agroecosystems. Crit. Rev. Plant Sci., 23, 175-193.
Weller D.M., 1988. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol., 26, 379-407.
Whipps J.M., 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot., 52, 487-511.
Whipps J.M., 1990. Carbon economy. In: Lynch J.M., ed. The rhizosphere. Chichester, UK: Wiley & Son, 59-87.
Yang C.-H. & Crowley D.E., 2000. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol., 66, 345-351.
To cite this article
About: Venant Nihorimbere*
University of Burundi. Faculty of Agricultural Sciences. PO Box 1550. Bujumbura (Burundi). E-mail: venant.nihorimbere@gmail.com – University of Burundi. Institut Supérieur d’Agriculture. BP 7268. Bujumbura (Burundi)
About: Marc Ongena*
Univ. Liège - Gembloux Agro-Bio Tech. Centre Wallon de Biologie Industrielle. Passage des Déportés, 2. B-5030 Gembloux (Belgique).
About: Maïté Smargiassi
Univ. Liège - Gembloux Agro-Bio Tech. Centre Wallon de Biologie Industrielle. Passage des Déportés, 2. B-5030 Gembloux (Belgique).
About: Philippe Thonart
Univ. Liège - Gembloux Agro-Bio Tech. Centre Wallon de Biologie Industrielle. Passage des Déportés, 2. B-5030 Gembloux (Belgique) – Univ. Liège. Service de Technologie microbienne. Boulevard du Rectorat, 29. B-4000 Liège (Belgique).






